

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.8 - phase: 0 /pseudo
(271 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
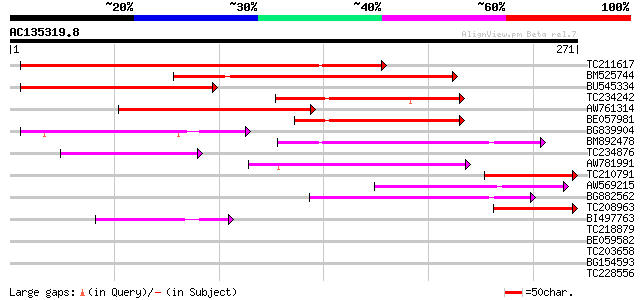
Score E
Sequences producing significant alignments: (bits) Value
TC211617 159 2e-39
BM525744 132 2e-31
BU545334 86 2e-17
TC234242 85 3e-17
AW761314 82 3e-16
BE057981 67 7e-12
BG839904 65 2e-11
BM892478 54 6e-08
TC234876 similar to UP|Q7RNR5 (Q7RNR5) 60S ribosomal protein L27... 52 4e-07
AW781991 50 8e-07
TC210791 45 3e-05
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 44 8e-05
BG882562 44 1e-04
TC208963 43 1e-04
BI497763 41 7e-04
TC218879 similar to UP|Q6ICW8 (Q6ICW8) At4g25180, partial (28%) 33 0.14
BE059582 32 0.40
TC203658 homologue to UP|PGMU_PEA (Q9SM60) Phosphoglucomutase, c... 29 2.6
BG154593 weakly similar to PIR|A86350|A86 F8K7.10 protein - Arab... 28 4.4
TC228556 weakly similar to UP|Q7PWJ2 (Q7PWJ2) ENSANGP00000021389... 28 4.4
>TC211617
Length = 534
Score = 159 bits (401), Expect = 2e-39
Identities = 77/175 (44%), Positives = 111/175 (63%)
Frame = -1
Query: 6 CERSGEHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVRYVAG 65
CERSG++K K + T +RKCGC FK+RG + W + ++ G+HNHE+ + +
Sbjct: 531 CERSGKYKCRKKEFVRRDTCTRKCGCPFKIRGKPMHGGEGWTVKLICGIHNHELAKTLVE 352
Query: 66 HLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNIKQVYNERHKFKKTKRGD 125
H AGRL +D+K I+ D+T S VKP+NI LK+ S T IKQ+YN R + + RGD
Sbjct: 351 HPYAGRLTDDEKNIIADMTKSNVKPRNIQLTLKEHNTNSCTTIKQIYNARSAYHSSIRGD 172
Query: 126 LTEMQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDST 180
TEMQ+L+ L+ + Y+H+ R K + V+D+FW HP +VKL NT V ++DST
Sbjct: 171 DTEMQHLMRLLKRDQYIHWHRLK-DQDVVRDLFWCHPDAVKLCNTCHLVFLIDST 10
>BM525744
Length = 411
Score = 132 bits (331), Expect = 2e-31
Identities = 61/136 (44%), Positives = 95/136 (69%)
Frame = +3
Query: 79 IVHDLTDSLVKPKNILTNLKKKRKESITNIKQVYNERHKFKKTKRGDLTEMQYLISKLEE 138
+V D+T +V+P+NIL LK + T I+ +YN R ++ +++G TEMQ+L+ LE
Sbjct: 6 LVIDMTKKMVEPRNILLTLKDHNND--TTIRHIYNARQAYRSSQKGPRTEMQHLLKLLEH 179
Query: 139 NGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTS 198
+ YV + R+ +S ++DIFW HP ++KL +F TVL +D+TYK N Y++PL EIVGVTS
Sbjct: 180 DQYVCWSRKVDDSDAIRDIFWAHPDAIKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGVTS 359
Query: 199 TYLTYSVDFSFMTAEK 214
T LT+SV F++M +++
Sbjct: 360 TELTFSVAFAYMESDE 407
>BU545334
Length = 556
Score = 85.9 bits (211), Expect = 2e-17
Identities = 41/94 (43%), Positives = 59/94 (62%)
Frame = -1
Query: 6 CERSGEHKLPKTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVRYVAG 65
CER+G++K A SRK GC FK+RG V W + ++ G+HNHE+ + +
Sbjct: 292 CERNGQYKCRNKXXVIRAIGSRKYGCPFKLRGKPVIGGQGWMMKLICGIHNHELAKSLVE 113
Query: 66 HLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKK 99
H AGRL +D+K I+ D+T S+VKP+NIL LK+
Sbjct: 112 HPYAGRLTKDEKTIIVDITKSMVKPRNILLILKE 11
>TC234242
Length = 478
Score = 85.1 bits (209), Expect = 3e-17
Identities = 43/95 (45%), Positives = 62/95 (65%), Gaps = 5/95 (5%)
Frame = +3
Query: 128 EMQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYR 187
EMQ+L+ LE + Y+H+ R K E V+DIFW HP SVKL N V ++DSTY+TN YR
Sbjct: 15 EMQHLMKFLERDQYIHWHRIKDED-VVRDIFWCHPDSVKLVNACNLVFLIDSTYRTNRYR 191
Query: 188 MPL-----FEIVGVTSTYLTYSVDFSFMTAEKEDN 217
+PL + VG+T T +T+S F+++ E+ +N
Sbjct: 192 LPLLDFVVLDFVGMTPTGMTFSASFAYVEGERVNN 296
>AW761314
Length = 331
Score = 82.0 bits (201), Expect = 3e-16
Identities = 41/94 (43%), Positives = 60/94 (63%)
Frame = +1
Query: 53 GVHNHEMVRYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNIKQVY 112
G NHE+V+ + H A RL +D+ I+ ++T S+VKPKNIL LK+ S T IKQ+Y
Sbjct: 49 GSDNHELVKSLVRHPYASRLTKDENIIITNMTKSMVKPKNILLTLKEHNVNSYTAIKQIY 228
Query: 113 NERHKFKKTKRGDLTEMQYLISKLEENGYVHYVR 146
N R + + RG T+MQ L+ LE++ Y+H+ R
Sbjct: 229 NARSAYCSSIRGSDTKMQQLMKLLEQDQYIHWHR 330
>BE057981
Length = 268
Score = 67.4 bits (163), Expect = 7e-12
Identities = 32/81 (39%), Positives = 51/81 (62%)
Frame = +2
Query: 137 EENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGV 196
E + Y+H+ R K E V+D+FW H +VKL N V ++++ YKTN YR+ L + GV
Sbjct: 14 ERDQYIHWHRLKDEF-VVRDLFWCHLDAVKLCNACHLVFLINNAYKTNRYRLLLLDFAGV 190
Query: 197 TSTYLTYSVDFSFMTAEKEDN 217
T +T+SV F+++ E+ +N
Sbjct: 191 TPARMTFSVGFAYLEGERVNN 253
>BG839904
Length = 712
Score = 65.5 bits (158), Expect = 2e-11
Identities = 35/117 (29%), Positives = 64/117 (53%), Gaps = 7/117 (5%)
Frame = -3
Query: 6 CERSGEHKLP-KTKVKHEATESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVRYVA 64
C+RSG+++ P K ++ + + +RKC C FK++G +++ W + ++ G HNH++ +
Sbjct: 338 CDRSGKYRGPYKNELSRKVSGTRKCECPFKLKGKALKKAEGWIVKVMCGCHNHDLXETLV 159
Query: 65 GHLLAGRLMEDDKKI------VHDLTDSLVKPKNILTNLKKKRKESITNIKQVYNER 115
GH AGRL ++K HD ++ + + K ++T IKQ+YN R
Sbjct: 158 GHPYAGRLSAEEKSFGRCFDKEHDEAX-----RHSIDH*KIITWVTVTTIKQIYNAR 3
>BM892478
Length = 429
Score = 54.3 bits (129), Expect = 6e-08
Identities = 31/128 (24%), Positives = 62/128 (48%)
Frame = +3
Query: 129 MQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 188
++Y S+ E+ Y E ++ +IFW S + F VL++D++Y+ +Y +
Sbjct: 33 LEYFQSRQAEDTGFFYAMEV-DNGNCMNIFWADGRSRYSCSHFGDVLVLDTSYRKTVYLV 209
Query: 189 PLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMM 248
P VGV + + + E E++FTW Q L+ + + +P V+ ++D ++
Sbjct: 210 PFATFVGVNHHKQPVLLGCALIADESEESFTWLFQTWLRAM--SGRLPLTVIADQDIAIQ 383
Query: 249 KAVTNALP 256
+A+ P
Sbjct: 384 RAIAKVFP 407
>TC234876 similar to UP|Q7RNR5 (Q7RNR5) 60S ribosomal protein L27 homolog,
partial (15%)
Length = 920
Score = 51.6 bits (122), Expect = 4e-07
Identities = 23/68 (33%), Positives = 37/68 (53%)
Frame = -3
Query: 25 ESRKCGCLFKVRGYVVRENNAWKLTILNGVHNHEMVRYVAGHLLAGRLMEDDKKIVHDLT 84
E+RKCGCLFK++ V W ++ HNH + + + H RL ++K I+ D+
Sbjct: 606 ENRKCGCLFKLQVKPVLGGEGWMKKLIRESHNHALAKLLVRHPYVDRLTINEKIIIGDMI 427
Query: 85 DSLVKPKN 92
++K KN
Sbjct: 426 KLIIKSKN 403
>AW781991
Length = 419
Score = 50.4 bits (119), Expect = 8e-07
Identities = 28/107 (26%), Positives = 48/107 (44%), Gaps = 1/107 (0%)
Frame = +3
Query: 115 RHKFKKTKRGDLT-EMQYLISKLEENGYVHYVREKPESQTVQDIFWTHPTSVKLFNTFPT 173
R+ ++ GD+ + YL EN Y + E Q++ ++FW P + + F
Sbjct: 84 RNNRLRSLEGDIQLVLDYLRQMHAENPNFFYAVQGDEDQSITNVFWADPKARMNYTFFGD 263
Query: 174 VLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTW 220
+ D+TY++N YR+P GV +F+ E E +F W
Sbjct: 264 TVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGCAFLINESEASFVW 404
>TC210791
Length = 456
Score = 45.4 bits (106), Expect = 3e-05
Identities = 20/44 (45%), Positives = 30/44 (67%)
Frame = +1
Query: 228 LLEPNSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
L+ +++MP V++T RD ++M AV P SS +LC FH+ KNV
Sbjct: 7 LIFKDNEMPPVIITVRDIALMDAVQVVFPSSSNLLCRFHISKNV 138
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 43.9 bits (102), Expect = 8e-05
Identities = 22/93 (23%), Positives = 48/93 (50%)
Frame = +1
Query: 175 LIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVDFSFMTAEKEDNFTWALQMLLKLLEPNSD 234
++ D+TY+ Y M L +GV + +T + + E +F+WAL+ L ++ +
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKA- 186
Query: 235 MPKVVVTNRDPSMMKAVTNALPDSSAILCYFHV 267
P+ ++T+ + + +A+ LP + C +H+
Sbjct: 187 -PQTILTDHNMWLKEAIAVELPQTKHAFCIWHI 282
>BG882562
Length = 416
Score = 43.5 bits (101), Expect = 1e-04
Identities = 25/108 (23%), Positives = 48/108 (44%)
Frame = +3
Query: 144 YVREKPESQTVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTY 203
YV + + ++++FW S ++ F V+ DST +N Y +PL VGV +
Sbjct: 90 YVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTCLSNNYEIPLVAFVGVNHHGKSV 269
Query: 204 SVDFSFMTAEKEDNFTWALQMLLKLLEPNSDMPKVVVTNRDPSMMKAV 251
+ + E + + W + L + P+ ++TN +M A+
Sbjct: 270 LLGCGLLADETFETYIWLFRAWLTCM--TGRPPQTIITNXCKAMQSAI 407
>TC208963
Length = 1163
Score = 43.1 bits (100), Expect = 1e-04
Identities = 19/40 (47%), Positives = 27/40 (67%)
Frame = +1
Query: 232 NSDMPKVVVTNRDPSMMKAVTNALPDSSAILCYFHVGKNV 271
+ DMP+V+VT D ++M AV P SS +LC FH+ +NV
Sbjct: 10 DDDMPQVIVTVGDIALMSAVQVVFPSSSNLLCRFHINQNV 129
>BI497763
Length = 421
Score = 40.8 bits (94), Expect = 7e-04
Identities = 20/66 (30%), Positives = 36/66 (54%)
Frame = +2
Query: 42 ENNAWKLTILNGVHNHEMVRYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKNILTNLKKKR 101
E W + +L G HNHE+ + + H GRL +D+K I+ ++T K TN+++ +
Sbjct: 35 EGKGWMVKLLCGSHNHELTKSLVEHPYVGRLTKDEKTIIVNMT------KKHFTNVERVQ 196
Query: 102 KESITN 107
+ + N
Sbjct: 197 CQWLYN 214
Score = 30.8 bits (68), Expect = 0.68
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = +3
Query: 87 LVKPKNILTNLKKKRKESITNIKQVYNERHK 117
L+ P+NIL LK+ T I+Q+YN R K
Sbjct: 153 LI*PRNILLMLKEYNANGCTTIRQIYNARGK 245
>TC218879 similar to UP|Q6ICW8 (Q6ICW8) At4g25180, partial (28%)
Length = 1277
Score = 33.1 bits (74), Expect = 0.14
Identities = 34/119 (28%), Positives = 50/119 (41%), Gaps = 20/119 (16%)
Frame = +1
Query: 9 SGEHKLPKTKVKHEATESRKCGCLFKVRGYVV--------RENNAWKLTILNGVHNHEMV 60
S + +LPK K T+ KC L +V + +EN AWKL ++ VH+H
Sbjct: 187 SKQSELPKPKRMISETKIAKCLELSRVDDMKILQNASLKSKENLAWKLHLVLEVHHH--- 357
Query: 61 RYVAGHLLAGRLMEDDKKIVHDLTDSLVKPKN------------ILTNLKKKRKESITN 107
+GH+ L + +VH L + P N +L +L K KES N
Sbjct: 358 -IPSGHMAQANL*IVAQAVVHHLN---ILPMNKFVQLLLRIKMMLLMHLPGKSKESTRN 522
>BE059582
Length = 285
Score = 31.6 bits (70), Expect = 0.40
Identities = 19/63 (30%), Positives = 31/63 (49%)
Frame = -3
Query: 71 RLMEDDKKIVHDLTDSLVKPKNILTNLKKKRKESITNIKQVYNERHKFKKTKRGDLTEMQ 130
RL+ D KK + +S+ P ++ K+KRK+ I NI ++ +R GD
Sbjct: 208 RLITDQKKKKKKVWESIFFPL*LMKEQKRKRKKEIKNIIKMLQKRV*LLAQVGGDFRSNS 29
Query: 131 YLI 133
YL+
Sbjct: 28 YLL 20
>TC203658 homologue to UP|PGMU_PEA (Q9SM60) Phosphoglucomutase, cytoplasmic
(Glucose phosphomutase) (PGM) , complete
Length = 2477
Score = 28.9 bits (63), Expect = 2.6
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = -3
Query: 86 SLVKPKNILTNLKKKRKESITNIKQVYNERHKFK 119
SL + N+ KKKRKE + IK+ Y H K
Sbjct: 2259 SLGEKSNLTAGTKKKRKERMLLIKRQYKRSHHIK 2158
>BG154593 weakly similar to PIR|A86350|A86 F8K7.10 protein - Arabidopsis
thaliana, partial (16%)
Length = 445
Score = 28.1 bits (61), Expect = 4.4
Identities = 13/27 (48%), Positives = 19/27 (70%)
Frame = +1
Query: 234 DMPKVVVTNRDPSMMKAVTNALPDSSA 260
+MP V V NRD S ++ V+NA+ S+A
Sbjct: 283 EMPGVYVVNRDGSNLRKVSNAMAFSTA 363
>TC228556 weakly similar to UP|Q7PWJ2 (Q7PWJ2) ENSANGP00000021389, partial
(6%)
Length = 897
Score = 28.1 bits (61), Expect = 4.4
Identities = 14/33 (42%), Positives = 15/33 (45%), Gaps = 4/33 (12%)
Frame = -3
Query: 133 ISKLEENGYVHYVREKPES----QTVQDIFWTH 161
I K+ GY HY R P S Q IFW H
Sbjct: 382 IQKITSEGYTHYSRNSPNSISYTQKYTCIFW*H 284
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,453,494
Number of Sequences: 63676
Number of extensions: 139665
Number of successful extensions: 811
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 799
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 807
length of query: 271
length of database: 12,639,632
effective HSP length: 96
effective length of query: 175
effective length of database: 6,526,736
effective search space: 1142178800
effective search space used: 1142178800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC135319.8


