

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.2 - phase: 0
(323 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
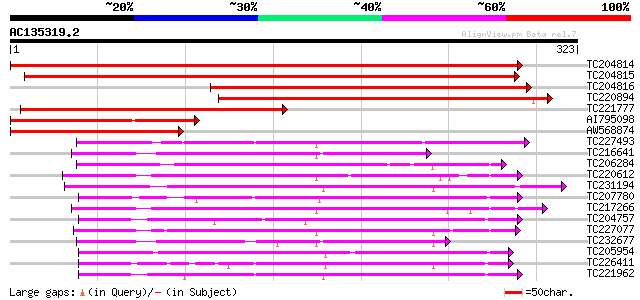
Score E
Sequences producing significant alignments: (bits) Value
TC204814 UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, complete 523 e-149
TC204815 similar to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein P... 475 e-134
TC204816 homologue to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein... 302 2e-82
TC220894 weakly similar to UP|Q8YLW4 (Q8YLW4) Oxidoreductase, pa... 226 1e-59
TC221777 similar to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein P... 211 3e-55
AI795098 homologue to GP|9622153|gb| seed maturation protein PM3... 169 2e-42
AW568874 similar to GP|9622153|gb|A seed maturation protein PM34... 154 5e-38
TC227493 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carri... 114 5e-26
TC216641 similar to UP|P93697 (P93697) CPRD12 protein, partial (... 102 2e-22
TC206284 similar to UP|Q8S305 (Q8S305) Short-chain alcohol dehyd... 101 4e-22
TC220612 weakly similar to UP|Q9SBM0 (Q9SBM0) Wts2L, partial (66%) 101 5e-22
TC231194 similar to UP|Q9LS70 (Q9LS70) Alcohol dehydrogenase-lik... 99 3e-21
TC207780 weakly similar to UP|Q91WL8 (Q91WL8) WW-domain oxidored... 97 1e-20
TC217266 similar to GB|AAL99238.1|22651517|AY082345 short-chain ... 96 3e-20
TC204757 similar to UP|Q9ASX2 (Q9ASX2) At1g07440/F22G5_16, parti... 95 5e-20
TC227077 weakly similar to UP|Q8H0D9 (Q8H0D9) Alcohol dehydroge,... 92 4e-19
TC232677 similar to UP|P93697 (P93697) CPRD12 protein, partial (... 91 5e-19
TC205954 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partia... 88 6e-18
TC226411 weakly similar to UP|TRNH_DATST (P50165) Tropinone redu... 87 8e-18
TC221962 similar to GB|BAA98195.1|8978342|AP002030 short chain a... 87 1e-17
>TC204814 UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, complete
Length = 1240
Score = 523 bits (1346), Expect = e-149
Identities = 248/292 (84%), Positives = 276/292 (93%)
Frame = +1
Query: 1 MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN 60
M +G QK PPQ+Q TQPGKEHAM P PQFT PDYKP+NKLQGKIA+VTGGDSGIGRAVCN
Sbjct: 91 MASGEQKFPPQQQQTQPGKEHAMTPVPQFTSPDYKPSNKLQGKIALVTGGDSGIGRAVCN 270
Query: 61 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEI 120
LFALEGATV FTYVKGHEDKDARDTL+M+K AKT++AKDPMAIP+DLG+DENCKRV+DE+
Sbjct: 271 LFALEGATVAFTYVKGHEDKDARDTLEMIKRAKTSDAKDPMAIPSDLGYDENCKRVVDEV 450
Query: 121 INAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNII 180
++AYGRIDILVNNAAEQYECG+VE+IDEPRLERVFRTNIFSYFFM RHALKHMKEGS+II
Sbjct: 451 VSAYGRIDILVNNAAEQYECGTVEDIDEPRLERVFRTNIFSYFFMARHALKHMKEGSSII 630
Query: 181 NTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFN 240
NTTSVNAYKG++ L+DYTSTKGAIVA+TR L+LQLVSKGIRVNGVAPGPIWTPLIPASF
Sbjct: 631 NTTSVNAYKGHAKLLDYTSTKGAIVAYTRGLALQLVSKGIRVNGVAPGPIWTPLIPASFK 810
Query: 241 EEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLIN 292
EE+TAQFG+ VPMKRAGQP+EVAPS+VFLASNQCSSYITGQVLHPNG ++N
Sbjct: 811 EEETAQFGAQVPMKRAGQPIEVAPSYVFLASNQCSSYITGQVLHPNGGTVVN 966
>TC204815 similar to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, partial
(98%)
Length = 1028
Score = 475 bits (1223), Expect = e-134
Identities = 227/282 (80%), Positives = 259/282 (91%)
Frame = +3
Query: 9 PPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGAT 68
PPQKQDTQPGKE+ MNP PQ+ YKP+NKLQGKIAVVTGGDSGIGRAVCNLF+LEGAT
Sbjct: 132 PPQKQDTQPGKEYLMNPPPQYNSSQYKPSNKLQGKIAVVTGGDSGIGRAVCNLFSLEGAT 311
Query: 69 VIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRID 128
VIFTYVKG ED+DA DTL+++K AKT +AKDP+AIP D+G++ENCK+V+DE+INAYGRID
Sbjct: 312 VIFTYVKGQEDRDASDTLEIIKKAKTEDAKDPLAIPVDVGYEENCKKVVDEVINAYGRID 491
Query: 129 ILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNIINTTSVNAY 188
ILVNNAAEQYE S+E+ID+ RLERVFRTNIFS+FFMT+HALKHMKEGS+IINTTSVNAY
Sbjct: 492 ILVNNAAEQYESDSLEDIDDARLERVFRTNIFSHFFMTKHALKHMKEGSSIINTTSVNAY 671
Query: 189 KGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFG 248
+G+ TL+DYTSTKGAIV FTRAL+LQLVSKGIRVNGVAPGPIWTPLI A+ NEE +FG
Sbjct: 672 QGDGTLVDYTSTKGAIVGFTRALALQLVSKGIRVNGVAPGPIWTPLIVATMNEETIVRFG 851
Query: 249 SDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDML 290
SDVPMKRAGQP+EVAPS+VFLASN CSSYITGQVLHPNG ++
Sbjct: 852 SDVPMKRAGQPIEVAPSYVFLASNICSSYITGQVLHPNGGII 977
>TC204816 homologue to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34,
partial (61%)
Length = 778
Score = 302 bits (773), Expect = 2e-82
Identities = 146/183 (79%), Positives = 162/183 (87%)
Frame = +1
Query: 115 RVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMK 174
RV E++NAYG ID VNNA YECG VE+ID RLERVFRTNIFSYFFMTRHALKHMK
Sbjct: 1 RVXXEVVNAYGCIDXXVNNAXXXYECGXVEDIDXXRLERVFRTNIFSYFFMTRHALKHMK 180
Query: 175 EGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPL 234
EGS+IINTTSVNAYKGN+ L+DYTSTKGAIVA+TR L+LQLVSKGIRVNGVAPGPIWTPL
Sbjct: 181 EGSSIINTTSVNAYKGNAKLLDYTSTKGAIVAYTRGLALQLVSKGIRVNGVAPGPIWTPL 360
Query: 235 IPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLINAS 294
IP+SF EE+TAQFG+ VPMKRAGQP+EVAPS+VFLA NQCSSYITGQVLHPNG ++N
Sbjct: 361 IPSSFKEEETAQFGAQVPMKRAGQPIEVAPSYVFLACNQCSSYITGQVLHPNGGTVVNG* 540
Query: 295 NVI 297
V+
Sbjct: 541 TVV 549
>TC220894 weakly similar to UP|Q8YLW4 (Q8YLW4) Oxidoreductase, partial (59%)
Length = 666
Score = 226 bits (576), Expect = 1e-59
Identities = 114/192 (59%), Positives = 140/192 (72%), Gaps = 2/192 (1%)
Frame = +1
Query: 120 IINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHMKEGSNI 179
++ Y D+ V NA EQ+ VEEI + +LERVF TNIFS FF+ +HALKHMKEGS I
Sbjct: 1 VVKEYXXXDVXVXNAXEQHLTXXVEEITQQQLERVFGTNIFSQFFLVKHALKHMKEGSCI 180
Query: 180 INTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASF 239
IN+TSVNAY GN +DYT+TKGAIVAFTR LS QL S+GIRVNGVAPGP+WTP+ PAS
Sbjct: 181 INSTSVNAYNGNPEALDYTATKGAIVAFTRGLSQQLASRGIRVNGVAPGPVWTPIQPASK 360
Query: 240 NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLINASNVI-- 297
E G +VPM R QP E+AP ++FLA+ Q SSY TGQVLHPNG M++NA ++
Sbjct: 361 PAEMIQNLGCEVPMNRVAQPCEIAPCYLFLATCQDSSYFTGQVLHPNGGMVVNA*AIVCL 540
Query: 298 YLFTHCMVFLTH 309
+LF +VFL +
Sbjct: 541 HLFPIELVFLAY 576
>TC221777 similar to UP|Q9LLQ6 (Q9LLQ6) Seed maturation protein PM34, partial
(45%)
Length = 517
Score = 211 bits (538), Expect = 3e-55
Identities = 104/152 (68%), Positives = 121/152 (79%)
Frame = +2
Query: 7 KIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEG 66
K P Q Q TQPGKEH MNP PQ T PD+K ANKLQGK+A+VTGGDS IGRAVC FA EG
Sbjct: 62 KFPAQSQKTQPGKEHVMNPLPQATNPDHKAANKLQGKVALVTGGDSRIGRAVCLCFAKEG 241
Query: 67 ATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGR 126
ATV FTYVKGHED+D DTL ML AKT+ A +P+AI AD+GFDENCK+VID ++ YGR
Sbjct: 242 ATVAFTYVKGHEDRDKDDTLKMLLEAKTSGADNPLAIAADIGFDENCKQVIDLVVKEYGR 421
Query: 127 IDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+D+LVNNAAEQ+ SVEEI + +LERVF T+
Sbjct: 422 LDVLVNNAAEQH*TNSVEEIAQQQLERVFSTH 517
>AI795098 homologue to GP|9622153|gb| seed maturation protein PM34 {Glycine
max}, partial (34%)
Length = 373
Score = 169 bits (427), Expect = 2e-42
Identities = 82/108 (75%), Positives = 93/108 (85%)
Frame = +1
Query: 1 MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN 60
M +G QK PPQ+Q TQPGKEHAM P PQFT PDYKP+NKLQGKIA+VTGGDSGIGRA CN
Sbjct: 52 MASGEQKFPPQQQQTQPGKEHAMTPVPQFTSPDYKPSNKLQGKIALVTGGDSGIGRAECN 231
Query: 61 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLG 108
LFA + + TYVKGHEDKDARDTL+M+K AKT++AKDPMAIP+DLG
Sbjct: 232 LFA*KVYRGL-TYVKGHEDKDARDTLEMIKRAKTSDAKDPMAIPSDLG 372
>AW568874 similar to GP|9622153|gb|A seed maturation protein PM34 {Glycine
max}, partial (33%)
Length = 308
Score = 154 bits (389), Expect = 5e-38
Identities = 71/99 (71%), Positives = 82/99 (82%)
Frame = +1
Query: 1 MTTGGQKIPPQKQDTQPGKEHAMNPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCN 60
M +G QK PPQ+Q TQPGKEHAM P PQFT PDYKP+NKLQ KIA+VT GDSGIG+ VCN
Sbjct: 10 MASGEQKFPPQQQQTQPGKEHAMTPVPQFTSPDYKPSNKLQRKIALVTEGDSGIGQTVCN 189
Query: 61 LFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
LF LE AT+ FTYVKGH+DKD ++TL M+K AKT+N KD
Sbjct: 190 LFTLESATMAFTYVKGHKDKDTKNTLKMIKRAKTSNTKD 306
>TC227493 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carrier-protein]
reductase, chloroplast precursor (3-ketoacyl-acyl
carrier protein reductase) , partial (83%)
Length = 1452
Score = 114 bits (286), Expect = 5e-26
Identities = 75/260 (28%), Positives = 133/260 (50%), Gaps = 2/260 (0%)
Frame = +1
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
K++ +AVVTG GIG+A+ G V+ Y + ++ + ++ K + +
Sbjct: 397 KVEAPVAVVTGASRGIGKAIALSLGKAGCKVLVNYARSSKEAE-----EVSKEIEEFGGQ 561
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+ D+ +++ + +I ++A+G +D+L+NNA + G + + + + + V N
Sbjct: 562 -ALTFGGDVSNEDDVESMIKTAVDAWGTVDVLINNAGITRD-GLLMRMKKSQWQDVIDLN 735
Query: 159 IFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLV 216
+ F T+ A K M K I+N SV GN +Y++ K ++ T+ ++ +
Sbjct: 736 LTGVFLCTQAAAKIMMKKRKGRIVNIASVVGLVGNVGQANYSAAKAGVIGLTKTVAKEYA 915
Query: 217 SKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSS 276
S+ I VN VAPG I + + A ++ + +P+ R GQP EVA FLA NQ +S
Sbjct: 916 SRNITVNAVAPGFIASDMT-AKLGQDIEKKILETIPLGRYGQPEEVAGLVEFLALNQAAS 1092
Query: 277 YITGQVLHPNGDMLINASNV 296
YITGQV +G M++ SN+
Sbjct: 1093YITGQVFTIDGGMVM*ISNL 1152
>TC216641 similar to UP|P93697 (P93697) CPRD12 protein, partial (74%)
Length = 1103
Score = 102 bits (255), Expect = 2e-22
Identities = 67/209 (32%), Positives = 107/209 (51%), Gaps = 4/209 (1%)
Frame = +3
Query: 36 PANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTA 95
P +L GK+A++TGG SG+G A LF+ GA V+ D D L ++
Sbjct: 291 PFRRLDGKVAIITGGASGLGAATARLFSKHGAYVVIA-----------DIQDDLGLSVAK 437
Query: 96 NAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLERV 154
+ + D+ +E+ + ++ ++ YG++DI+ NNA E S+ + ++ ERV
Sbjct: 438 ELESASYVHCDVTKEEDVENCVNTTVSKYGKLDIMFNNAGVSDEIKTSILDNNKSDFERV 617
Query: 155 FRTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRAL 211
N+ F T+HA + M K+G IINT SV G YTS+K A++ T+
Sbjct: 618 ISVNLVGPFLGTKHAARVMIPAKKGC-IINTASVAGCIGGGATHAYTSSKHALIGLTKNT 794
Query: 212 SLQLVSKGIRVNGVAPGPIWTPLIPASFN 240
+++L GIRVN ++P + TPL FN
Sbjct: 795 AVELGQHGIRVNWLSPYLVVTPLSKKYFN 881
>TC206284 similar to UP|Q8S305 (Q8S305) Short-chain alcohol dehydrogenase
(Fragment), partial (94%)
Length = 1100
Score = 101 bits (252), Expect = 4e-22
Identities = 66/248 (26%), Positives = 128/248 (51%), Gaps = 3/248 (1%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+ +GK+A+VT GIG A+ LEGA+V+ + K A + L A
Sbjct: 86 RFEGKVAIVTASTQGIGLAIAERLGLEGASVVISSRKQQNVDAAAEQL-------RAKGI 244
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
+ + + + K +ID+ + YG+ID++V+NAA ++ + + L++++ N
Sbjct: 245 QVLGVVCHVSSAQQRKNLIDKTVQKYGKIDVVVSNAAANPSVDAILQTKDSVLDKLWEIN 424
Query: 159 IFSYFFMTRHALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSK 218
+ + + + A+ H+++GS+++ +S+ + +L Y TK A++ T+AL+ ++ +
Sbjct: 425 VKATILLLKDAVPHLQKGSSVVIISSIAGFNPPPSLAMYGVTKTALLGLTKALAAEM-AP 601
Query: 219 GIRVNGVAPGPIWTPLIPASF---NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCS 275
RVN VAPG + P ASF N+ + + R G ++ + FLAS+ +
Sbjct: 602 NTRVNCVAPG--FVPTNFASFITSNDAVKKELEEKTLLGRLGTTEDMGAAAAFLASDD-A 772
Query: 276 SYITGQVL 283
+YITG+ +
Sbjct: 773 AYITGETI 796
>TC220612 weakly similar to UP|Q9SBM0 (Q9SBM0) Wts2L, partial (66%)
Length = 1130
Score = 101 bits (251), Expect = 5e-22
Identities = 79/274 (28%), Positives = 135/274 (48%), Gaps = 12/274 (4%)
Frame = +3
Query: 31 CPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLK 90
C D + +L+ K+A++TGG SGIG A LF GA V+ D +D L
Sbjct: 126 CSDAPLSKRLEDKVALITGGASGIGEATARLFLRHGAKVVIA--------DIQDNLGHSL 281
Query: 91 MAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEP 149
+ + + D+ D + + + ++ +G++DIL +NA S+ +D
Sbjct: 282 CQNLNSGNNISYVHCDVTNDNDVQIAVKAAVSRHGKLDILFSNAGIGGNSDSSIIALDPA 461
Query: 150 RLERVFRTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVA 206
L+RVF N+F F+ +HA + M K GS + +++V+ S YT++K A+V
Sbjct: 462 DLKRVFEVNVFGAFYAAKHAAEIMIPRKIGSIVFTSSAVSVTHPGSP-HPYTASKYAVVG 638
Query: 207 FTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKT------AQFGS--DVPMKRAGQ 258
+ L ++L GIRVN ++P + TPL+ EK A+ G+ V +K
Sbjct: 639 LMKNLCVELGKHGIRVNCISPYAVATPLLTRGMGMEKEMVEELFAEAGNLKGVVLKEE-- 812
Query: 259 PVEVAPSFVFLASNQCSSYITGQVLHPNGDMLIN 292
++A + +FLAS++ S Y++G L +G +N
Sbjct: 813 --DLAEAALFLASDE-SKYVSGVNLVVDGGYSVN 905
>TC231194 similar to UP|Q9LS70 (Q9LS70) Alcohol dehydrogenase-like protein,
partial (58%)
Length = 1197
Score = 98.6 bits (244), Expect = 3e-21
Identities = 78/293 (26%), Positives = 139/293 (46%), Gaps = 7/293 (2%)
Frame = +2
Query: 32 PDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKM 91
P K +++L+GK+A++TG SGIG LFA GA ++ T ++ + ++
Sbjct: 11 PMTKQSSRLEGKVALITGAASGIGEETVRLFAEHGALIVATDIQDEQGH---------RV 163
Query: 92 AKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRL 151
A + ++ D+ + + I+ + +GRID+L +NA + ++D
Sbjct: 164 AASIGSERVTYHHCDVRDENQVEETINFTLEKHGRIDVLFSNAGVIGSLSGILDLDLNEF 343
Query: 152 ERVFRTNIFSYFFMTRHALKHMKEGS---NIINTTSVNAYKGNSTLIDYTSTKGAIVAFT 208
+ TN+ +H + M S +II TTSV A G + YT++K A++
Sbjct: 344 DNTMATNVRGVAATIKHTARAMVAKSTRGSIICTTSVAATIGGTGPHGYTTSKHALLGLV 523
Query: 209 RALSLQLVSKGIRVNGVAPGPIWTPLIPASFN---EEKTAQFGSDVPMKRAG-QPVEVAP 264
++ +L + GIRVN ++P + TPL +FN E+ A S +K + +A
Sbjct: 524 KSACSELGAYGIRVNSISPFGVATPLACKAFNFEPEQVEANSCSQANLKGVVLKARHIAE 703
Query: 265 SFVFLASNQCSSYITGQVLHPNGDMLINASNVIYLFTHCMVFLTHSFHFYFSG 317
+ +FLAS+ + YI+G L +G + N Y FT + + H + +G
Sbjct: 704 AALFLASDDAAVYISGHNLVVDGG--FSVVNRSYSFTPA*LHVEPKKHSFVAG 856
>TC207780 weakly similar to UP|Q91WL8 (Q91WL8) WW-domain oxidoreductase (Mus
musculus 12 days embryo female mullerian duct includes
surrounding region cDNA, RIKEN full-length enriched
library, clone:6820434G18 product:WW domain-containing
oxidoreductase, full insert sequence), partial (7%)
Length = 1054
Score = 96.7 bits (239), Expect = 1e-20
Identities = 78/270 (28%), Positives = 134/270 (48%), Gaps = 17/270 (6%)
Frame = +1
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L+GK+A++TGG SGIG + F GA+V + G + + + +L+
Sbjct: 55 LKGKVALITGGASGIGFEISTQFGKHGASVA---LMGRRKQVLQSAVSVLQ--------- 198
Query: 100 PMAIPA-----DLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERV 154
+ IPA D+ E+ RV++ +GRIDILVN AA + S E++ V
Sbjct: 199 SLVIPAVGFEGDVRKQEDAARVVESTFKHFGRIDILVNAAAGNF-LVSAEDLSSNGFRTV 375
Query: 155 FRTNIFSYFFMTRHALKHMKE----------GSNIINTTSVNAYKGNSTLIDYTSTKGAI 204
+ F M ALK++K+ G +IIN ++ Y + I ++ K A+
Sbjct: 376 LDIDSVGTFTMCHEALKYLKKGGEGRSNSSSGGSIINISATLHYTASWYQIHVSAAKAAV 555
Query: 205 VAFTRALSLQL-VSKGIRVNGVAPGPIW-TPLIPASFNEEKTAQFGSDVPMKRAGQPVEV 262
A TR L+L+ IRVNG+APGPI TP + +E +++ +P+ + G+ ++
Sbjct: 556 DATTRNLALEWGTDYDIRVNGIAPGPISDTPGMSKLAPDEISSKARDYMPLYKLGEKWDI 735
Query: 263 APSFVFLASNQCSSYITGQVLHPNGDMLIN 292
A + +FL S+ +I G ++ +G + ++
Sbjct: 736 AMAALFLVSD-AGKFINGDIMIVDGGLWLS 822
>TC217266 similar to GB|AAL99238.1|22651517|AY082345 short-chain
dehydrogenase/reductase {Arabidopsis thaliana;} ,
partial (93%)
Length = 1133
Score = 95.5 bits (236), Expect = 3e-20
Identities = 80/284 (28%), Positives = 134/284 (47%), Gaps = 13/284 (4%)
Frame = +2
Query: 36 PANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTA 95
P +L GK+A+VTGG SGIG ++ LF + GA + D +D L
Sbjct: 56 PTQRLLGKVALVTGGASGIGESIVRLFHIHGAKICIA--------DVQDNLGKQVCQSLG 211
Query: 96 NAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERV 154
+ + + + D+ +++ +D + +G + I+VNNA C + D ++V
Sbjct: 212 DEANVVFVHCDVTVEDDVSHAVDFTVGKFGTLHIIVNNAGISGSPCSDIRNADLSEFDKV 391
Query: 155 FRTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALS 212
F N F +HA + M K+ +II+ SV + G YT +K A++ T+ ++
Sbjct: 392 FSVNTKGVFHGMKHAARIMIPKKKGSIISLCSVASAIGGLGPHAYTGSKYAVLGLTKNVA 571
Query: 213 LQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFG----SDVPMKRAG-QPVE-----V 262
+L IRVN V+P + T L A E++ D + A Q VE V
Sbjct: 572 AELGKHAIRVNCVSPYGVATGLALAHLPEDERTDDALVSFRDFTGRMANLQGVELTTHDV 751
Query: 263 APSFVFLASNQCSSYITGQVLHPNGDMLINASNVIYLFTHCMVF 306
A + +FLAS+ + YI+G+ L +G +A++ + +F MVF
Sbjct: 752 ANAVLFLASDD-AKYISGENLMVDGG-FTSANHSLQVFR*LMVF 877
>TC204757 similar to UP|Q9ASX2 (Q9ASX2) At1g07440/F22G5_16, partial (90%)
Length = 1257
Score = 94.7 bits (234), Expect = 5e-20
Identities = 80/259 (30%), Positives = 120/259 (45%), Gaps = 6/259 (2%)
Frame = +1
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
LQG A+VTGG GIG A+ A GATV AR+ ++ + N K
Sbjct: 97 LQGMTALVTGGSKGIGYAIVEELAQLGATV---------HTCARNEAELNESLNEWNTKG 249
Query: 100 PMAIPADLGFDENCKR--VIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFR 156
+ +R +I + N + G+++ILVNN +++ + E +
Sbjct: 250 YRVTGSVCDVASRAERQDLIARVSNEFNGKLNILVNNVGTNVPKHTLD-VTEEDFSFLIN 426
Query: 157 TNIFSYFFMTR--HALKHMKEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
TN+ S + +++ H L E +NII +S+ Y +TKGA+ T+ L+ +
Sbjct: 427 TNLESAYHLSQLAHPLLKASEAANIIFISSIAGVLSIGIGSTYGATKGAMNQLTKNLACE 606
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQ-FGSDVPMKRAGQPVEVAPSFVFLASNQ 273
IR N VAPGPI TPL F EK F S P+ R G+ EV+ FL
Sbjct: 607 WAKDNIRTNCVAPGPIKTPLGDKHFKNEKLLNAFISQTPLGRIGEAEEVSSLVAFLCL-P 783
Query: 274 CSSYITGQVLHPNGDMLIN 292
+SYITGQ + +G + +N
Sbjct: 784 AASYITGQTICVDGGLTVN 840
>TC227077 weakly similar to UP|Q8H0D9 (Q8H0D9) Alcohol dehydroge, partial
(88%)
Length = 1041
Score = 91.7 bits (226), Expect = 4e-19
Identities = 72/260 (27%), Positives = 126/260 (47%), Gaps = 5/260 (1%)
Frame = +2
Query: 37 ANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTAN 96
A +L+GK+A++TGG SGIG LF+ GA V+ D +D L + K +
Sbjct: 158 ARRLEGKVALITGGASGIGECTARLFSKHGAKVVIA--------DIQDELGH-SICKDLD 310
Query: 97 AKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLERVF 155
+ I D+ +EN + ++ ++ YG++DI+ ++A S+ + E+V
Sbjct: 311 SSSATYIHCDVTKEENIEHAVNTTVSKYGKLDIMHSSAGIVGAWNPSILHNKKSHFEQVI 490
Query: 156 RTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSL 213
N+ F +HA + M +I+ S+ G YTS+K IV R ++
Sbjct: 491 SVNLVGTFLGIKHAARVMIPSGRGSIVAMASICGRIGGVASHAYTSSKHGIVGLVRNTAV 670
Query: 214 QLVSKGIRVNGVAPGPIWTPLIPASFN--EEKTAQFGSDVPMKRAGQPVEVAPSFVFLAS 271
+L + GIRVN V+P + TP+ N +E A S++ +P +VA + ++L S
Sbjct: 671 ELGTLGIRVNSVSPYAVPTPMSKTFLNTDDEGIAALYSNL-KGTVLKPQDVAEAVLYLGS 847
Query: 272 NQCSSYITGQVLHPNGDMLI 291
++ S Y++G L +G +
Sbjct: 848 DE-SKYVSGHDLVVDGGFTV 904
>TC232677 similar to UP|P93697 (P93697) CPRD12 protein, partial (83%)
Length = 766
Score = 91.3 bits (225), Expect = 5e-19
Identities = 68/225 (30%), Positives = 110/225 (48%), Gaps = 12/225 (5%)
Frame = +1
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+A+++GG SGIG A LF+ GA V+ D D L ++ + +
Sbjct: 103 RLEGKVALISGGASGIGEATARLFSKHGAHVVIA-----------DIQDDLGLSLCKHLE 249
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRL------- 151
+ D+ + + + ++ I+ YG +DI+ NN G ++EI L
Sbjct: 250 SASYVHCDVTNENDVQNAVNTAISKYGNLDIMFNNP------GIIDEIKTSILDNSKFDS 411
Query: 152 ERVFRTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFT 208
ERV N+ F T+HA + M K GS IINT SV YTS+K A++
Sbjct: 412 ERVISVNLVGPFLGTKHAARVMIPAKRGS-IINTASVAGTFSGGASHAYTSSKHALIGLM 588
Query: 209 RALSLQLVSKGIRVNGVAPGPIWTPLIPASFN--EEKTAQFGSDV 251
+ +++L GIRVN ++P + TPL FN E++ + S++
Sbjct: 589 KNTAVELGQFGIRVNCLSPYVVATPLTKKCFNLDEDRNGEIYSNL 723
>TC205954 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partial (89%)
Length = 958
Score = 87.8 bits (216), Expect = 6e-18
Identities = 70/254 (27%), Positives = 109/254 (42%), Gaps = 6/254 (2%)
Frame = +1
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
LQ ++A+VTG GIGR + A GA ++ Y D+ +
Sbjct: 82 LQDRVAIVTGSSRGIGREIALHLASLGARLVVNYTSNSAQADS--VAAQINAGSATTTPR 255
Query: 100 PMAIPADLGFDENCKRVIDEIINAYGR-IDILVNNAAE-QYECGSVEEIDEPRLERVFRT 157
+ + AD+ K + D A+ I ILVN+A SV + +R F
Sbjct: 256 AVVVQADVSDPAQVKSLFDSAERAFDSPIHILVNSAGVIDGTYPSVADTTVESFDRTFAV 435
Query: 158 NIFSYFFMTRHALKHMKEGSN----IINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSL 213
N F R A +K G ++ T+ V A + Y ++K A+ A + L+
Sbjct: 436 NARGAFACAREAANRLKRGGGGRIILLTTSQVVALRPGYGA--YAASKAAVEAMVKILAK 609
Query: 214 QLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQ 273
+L I N VAPGPI T + EE + + P+ R G+ +VAP FLA++
Sbjct: 610 ELKGTQITANCVAPGPIATEMFFEGKTEEVVNRIVQESPLGRLGETKDVAPVVGFLATD- 786
Query: 274 CSSYITGQVLHPNG 287
S ++ GQ++ NG
Sbjct: 787 ASEWVNGQIVRVNG 828
>TC226411 weakly similar to UP|TRNH_DATST (P50165) Tropinone reductase
homolog (P29X) , partial (83%)
Length = 1109
Score = 87.4 bits (215), Expect = 8e-18
Identities = 79/256 (30%), Positives = 121/256 (46%), Gaps = 8/256 (3%)
Frame = +3
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
L G A+VTGG GIG A+ A GATV ++ D L+ K +K N
Sbjct: 93 LHGMTALVTGGTRGIGHAIVEELAEFGATV---HICARNQDDIDKCLEEWK-SKGLNVTG 260
Query: 100 PMAIPADLGFDENCKRVIDEIINA--YGRIDILVNNAAEQYECGSVEEIDEPRLERVFRT 157
+ DL + KR++ EI+ + +G+++ILVNNAA + + + + T
Sbjct: 261 SVC---DLLCSDQRKRLM-EIVGSIFHGKLNILVNNAATNIT-KKITDYTAEDISAIMGT 425
Query: 158 NIFSYFFMTRHALKHMKEGSN--IINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQL 215
N S + + + A +K+ N I+ +SV K Y ++KGA+ FT+ L+L+
Sbjct: 426 NFESVYHLCQVAHPLLKDSGNGSIVFISSVAGLKALPVFSVYAASKGAMNQFTKNLALEW 605
Query: 216 VSKGIRVNGVAPGPIWTPLIPASFN----EEKTAQFGSDVPMKRAGQPVEVAPSFVFLAS 271
IR N VAPGP+ T L+ N E S + R G+ E++ FL
Sbjct: 606 AKDNIRANAVAPGPVKTKLLECIVNSSEGNESINGVVSQTFVGRMGETKEISALVAFLCL 785
Query: 272 NQCSSYITGQVLHPNG 287
+SYITGQV+ +G
Sbjct: 786 -PAASYITGQVICVDG 830
>TC221962 similar to GB|BAA98195.1|8978342|AP002030 short chain alcohol
dehydrogenase-like {Arabidopsis thaliana;} , partial
(91%)
Length = 1245
Score = 87.0 bits (214), Expect = 1e-17
Identities = 74/259 (28%), Positives = 118/259 (44%), Gaps = 6/259 (2%)
Frame = +1
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK- 98
LQG A+VTGG GIG A+ GA V AR+ D+ K K N
Sbjct: 292 LQGMTALVTGGTRGIGHAIVEELTGFGARV---------HTCARNEHDLTKCLKNWNDSG 444
Query: 99 -DPMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFR 156
D D+ + +++ + + + G+++IL+NN V + +
Sbjct: 445 FDVTGSVCDVSVPHQREALMESVSSLFHGKLNILINNVGTNIR-KPVTDFTSAEFSTLID 621
Query: 157 TNIFSYFFMTRHALKHMKEGS--NIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
TN+ S F + + A +K +++ +SV+ + ++ +TKGAI TR L+ +
Sbjct: 622 TNLGSVFHLCQLAYPLLKASGMGSVVFVSSVSGFVSLKSMSVQGATKGAINQLTRNLACE 801
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASF-NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQ 273
IR N VAP I T L+ N++ + S P++R G P EV+ FL
Sbjct: 802 WAKDNIRSNAVAPWYIKTSLVEQVLSNKDYLEEVYSRTPLRRLGDPAEVSSLVAFLCL-P 978
Query: 274 CSSYITGQVLHPNGDMLIN 292
SSYITGQ++ +G M +N
Sbjct: 979 ASSYITGQIICIDGGMSVN 1035
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,343,726
Number of Sequences: 63676
Number of extensions: 177325
Number of successful extensions: 1114
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 1055
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1060
length of query: 323
length of database: 12,639,632
effective HSP length: 97
effective length of query: 226
effective length of database: 6,463,060
effective search space: 1460651560
effective search space used: 1460651560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135319.2


