

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
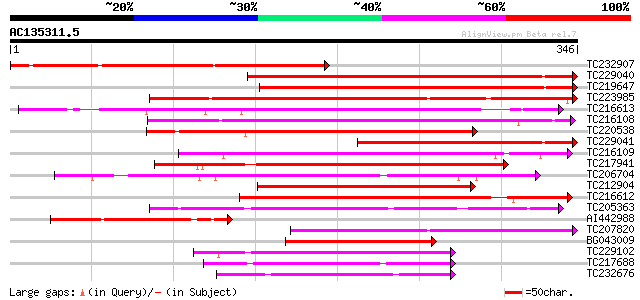
Query= AC135311.5 + phase: 0
(346 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232907 265 2e-71
TC229040 weakly similar to GB|AAP68276.1|31711840|BT008837 At3g6... 265 3e-71
TC219647 263 9e-71
TC223985 similar to UP|O04696 (O04696) DNA-binding protein PD1, ... 223 8e-59
TC216613 homologue to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, p... 206 1e-53
TC216108 201 4e-52
TC220538 similar to UP|Q9FHM5 (Q9FHM5) Similarity to DNA-binding... 186 1e-47
TC229041 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, par... 182 1e-46
TC216109 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, par... 178 4e-45
TC217941 similar to UP|Q9FIR1 (Q9FIR1) Similarity to AT-hook DNA... 174 7e-44
TC206704 similar to UP|Q94F52 (Q94F52) At2g33620/F4P9.39, partia... 164 7e-41
TC212904 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, par... 158 4e-39
TC216612 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, par... 157 5e-39
TC205363 similar to UP|Q9ZRR7 (Q9ZRR7) SAP1 protein, partial (59%) 153 1e-37
AI442988 similar to GP|2213534|emb| DNA-binding PD1-like protein... 120 9e-28
TC207820 similar to UP|O04696 (O04696) DNA-binding protein PD1, ... 120 9e-28
BG043009 weakly similar to GP|17979485|gb| AT4g12080/F16J13_150 ... 119 2e-27
TC229102 similar to UP|Q9LTA2 (Q9LTA2) Similarity to AT-hook DNA... 104 5e-23
TC217688 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like,... 98 5e-21
TC232676 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like,... 94 9e-20
>TC232907
Length = 762
Score = 265 bits (678), Expect = 2e-71
Identities = 137/195 (70%), Positives = 156/195 (79%)
Frame = +3
Query: 1 MDHGDHMALSNSASYYMQQRVLPGSGAQPELHVSPSFNQLSNPNLPFQSNIGGGGSNIGT 60
MD GD MALS S YYMQQR +PGSGA PELH+SP+ +SNPNLPFQS+IGGG IG+
Sbjct: 192 MDRGDQMALSGS--YYMQQRGIPGSGAPPELHISPNMRPISNPNLPFQSSIGGG--TIGS 359
Query: 61 TLPLESSAISSQGVNMSGHTGVPSGETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGT 120
TLPLESSAIS+ GVN+ TG P GE VKRKRGRPRKYG D VSLAL+P+PT SS PG
Sbjct: 360 TLPLESSAISAHGVNVGAPTGAPPGEPVKRKRGRPRKYGTDGSVSLALTPTPTSSSYPGA 539
Query: 121 MTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQV 180
+TQ KRG GRPPG+GKKQQLAS GELMSGSAG GF PH+I IASGEDI KI+ FSQ
Sbjct: 540 LTQS-QKRGIGRPPGTGKKQQLASLGELMSGSAGMGFTPHIINIASGEDITTKIMAFSQQ 716
Query: 181 RARALCVLSSSGSVS 195
ARA+C+LS++G+VS
Sbjct: 717 GARAVCILSANGAVS 761
>TC229040 weakly similar to GB|AAP68276.1|31711840|BT008837 At3g61310
{Arabidopsis thaliana;} , partial (42%)
Length = 893
Score = 265 bits (676), Expect = 3e-71
Identities = 131/201 (65%), Positives = 157/201 (77%)
Frame = +3
Query: 146 GELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSIS 205
GELMSGSAG GF PH+I IA GEDIA KI+ FSQ RA+C+LS++G+VS+V +R+PS S
Sbjct: 6 GELMSGSAGMGFTPHIITIAVGEDIATKIMAFSQQGPRAICILSANGAVSTVTLRQPSTS 185
Query: 206 GGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPV 265
GGT+ YEG F I+ +SG Y+ ++G +RNR LS+SL PDGR+ GG VGG L+AASPV
Sbjct: 186 GGTVTYEGRFEIVCLSGSYLVADSGGTRNRTVALSVSLASPDGRVIGGGVGGVLIAASPV 365
Query: 266 QVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPTSSLSPW 325
QV++GSF WG K K KKKE SE AE +E+DHQ HNP A+NSISPNQNLTPTSSLSPW
Sbjct: 366 QVILGSFSWGASKTKIKKKEGSEGAEVAMETDHQTVHNPVAVNSISPNQNLTPTSSLSPW 545
Query: 326 SAASRQMDMGNSHADIDLMRG 346
ASR +DM NSH DIDLMRG
Sbjct: 546 -PASRPLDMRNSHIDIDLMRG 605
>TC219647
Length = 1007
Score = 263 bits (672), Expect = 9e-71
Identities = 131/195 (67%), Positives = 153/195 (78%), Gaps = 1/195 (0%)
Frame = +1
Query: 153 AGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYE 212
AG GF PH+I IASGEDIA KI+ F Q R +C+LS++G+VS+V +R+PS SGGT+ YE
Sbjct: 1 AGMGFTPHIINIASGEDIATKIMAFXQQGPRVVCILSANGAVSTVTLRQPSTSGGTVTYE 180
Query: 213 GHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF 272
G F I+ +SG Y+ TENG SRNR GGLS+SL PDGR+ GG VGG L+A+SPVQV++GSF
Sbjct: 181 GRFEIVCLSGSYLVTENGGSRNRTGGLSVSLASPDGRVIGGGVGGVLIASSPVQVVVGSF 360
Query: 273 LWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPT-SSLSPWSAASRQ 331
LWG K KNKKKESSE AE VESDHQG HNP +LNSIS NQNL PT SLSPWS SR
Sbjct: 361 LWGGSKTKNKKKESSEGAEVAVESDHQGVHNPVSLNSISQNQNLPPTPPSLSPWS-TSRP 537
Query: 332 MDMGNSHADIDLMRG 346
+DM NSH DIDLMRG
Sbjct: 538 LDMRNSHVDIDLMRG 582
>TC223985 similar to UP|O04696 (O04696) DNA-binding protein PD1, partial
(59%)
Length = 901
Score = 223 bits (569), Expect = 8e-59
Identities = 127/265 (47%), Positives = 174/265 (64%), Gaps = 4/265 (1%)
Frame = +1
Query: 86 ETVKRKRGRPRKYGADRVVSLALSP-SPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLAS 144
E VK+KRGRPRKYG D VSL LSP S T SS PG+ T KR RGRPPGSG+KQQLA+
Sbjct: 1 EPVKKKRGRPRKYGPDGSVSLMLSPMSATASSTPGSGT-SSEKRPRGRPPGSGRKQQLAT 177
Query: 145 FGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSI 204
GE M+ SAG F PHVI + EDI AK+L+F++ R RA+C+L+ +G++SSV +R+P+
Sbjct: 178 LGEWMNSSAGLAFSPHVITVGVDEDIVAKLLSFARQRPRAVCILTGTGTISSVTLRQPAS 357
Query: 205 SGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASP 264
+ + YEG F I+ +SG Y+ E G NR GG+S+SL PDG + GG V LVA+SP
Sbjct: 358 TSIGVTYEGRFQILCLSGSYLVAEEGGPHNRTGGMSVSLSSPDGHIIGGGV-TRLVASSP 534
Query: 265 VQVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNSI-SPNQNLTPTSSLS 323
VQV+ SF++G K K K+ ++ + S+ Q + A+ S+ PNQN T + +
Sbjct: 535 VQVVACSFVYGGSKPKTKQVTTTTTKD--TSSEPQSSDKLASPGSVPPPNQNYTSSPAPG 708
Query: 324 PWSAASRQMDMGNSHA--DIDLMRG 346
W A+SR +++ ++HA IDL RG
Sbjct: 709 IWPASSRPVEVKSAHAHTGIDLTRG 783
>TC216613 homologue to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, partial (44%)
Length = 1790
Score = 206 bits (524), Expect = 1e-53
Identities = 129/342 (37%), Positives = 188/342 (54%), Gaps = 9/342 (2%)
Frame = +2
Query: 6 HMALSNSASYYMQQRV-LPGSGAQPELHVSPSFNQLSNPNLPFQSNIGGGGSNIGTTLPL 64
H L+ AS + V + GS A + HV+P + NP G+T +
Sbjct: 530 HKCLNMEASGGVSSGVTVVGSDAPSDYHVAP---RTDNP-----------APASGSTTQI 667
Query: 65 ESSAISSQGVNMSGHTGV----PSGETVKRKRGRPRKYGADRVVSLALSPSPTPSSNP-- 118
++A S+ + HT P+ K+KRGRPRKY D V++ALSP P SS P
Sbjct: 668 PATAGSALSPSHPPHTAAMEAYPATMPAKKKRGRPRKYAPDGSVTMALSPKPISSSAPLP 847
Query: 119 GTMTQGGPKRGRGRPPGSGKKQ--QLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILT 176
+ KRG+ +P S K +L + GE ++ S G F PH+I + SGED+ K+++
Sbjct: 848 PVIDFSSEKRGKIKPTSSVSKAKFELENLGEWVACSVGANFTPHIITVNSGEDVTMKVIS 1027
Query: 177 FSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRD 236
FSQ RA+C+LS++G +SSV +R+P SGGTL YEG F I+S+SG ++P E+G +R+R
Sbjct: 1028FSQQGPRAICILSANGVISSVTLRQPDSSGGTLTYEGRFEILSLSGSFMPNESGGTRSRS 1207
Query: 237 GGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSEDAEGTVES 296
GG+S+SL PDGR+ GG V G LVAASPVQV++GSFL G + +K+ E
Sbjct: 1208GGMSVSLASPDGRVVGGGVAGLLVAASPVQVVVGSFLAGNQHEQKPRKQKHE-------- 1363
Query: 297 DHQGAHNPAALNSISPNQNLTPTSSLSPWSAASRQMDMGNSH 338
++S++P + P S+L P S S + N +
Sbjct: 1364---------VISSVTP-AAVVPISTLDPVSILSAASSIRNDN 1459
>TC216108
Length = 1568
Score = 201 bits (511), Expect = 4e-52
Identities = 116/271 (42%), Positives = 157/271 (57%), Gaps = 10/271 (3%)
Frame = +1
Query: 85 GETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLAS 144
G K+KRGRPRKYG D + S+ALSP P SS P K+ RG+P K
Sbjct: 397 GTAAKKKRGRPRKYGPDGLNSMALSPIPISSSAPFANEFSSGKQ-RGKPRAMEYKLPKKV 573
Query: 145 FGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSI 204
+L S GT F+PH+I + +GEDI K+++FSQ RA+C+LS+SG +S+V +R+P
Sbjct: 574 GVDLFGDSVGTNFMPHIIAVNTGEDITMKVISFSQQGPRAICILSASGVISNVTLRQPDS 753
Query: 205 SGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASP 264
SGGTL YEG F I+S+SG ++PT+N +R+R GG+S+SL PDGR+ GG V G LVAA P
Sbjct: 754 SGGTLTYEGRFEILSLSGSFMPTDNQGTRSRSGGMSVSLSSPDGRIVGGGVAGLLVAAGP 933
Query: 265 VQVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNS----------ISPNQ 314
VQV++GSFL + K KK S+ A V + P N + N
Sbjct: 934 VQVVVGSFLPNNPQDKKPKKPKSDYAPANVTPSIAVSSAPPPTNGEKEDVMGGHLLHNNS 1113
Query: 315 NLTPTSSLSPWSAASRQMDMGNSHADIDLMR 345
T S+ SP SA R+ + N H+ D M+
Sbjct: 1114GTTLNSNFSPPSAFRRE-NWVNMHSMADSMK 1203
>TC220538 similar to UP|Q9FHM5 (Q9FHM5) Similarity to DNA-binding protein,
partial (41%)
Length = 825
Score = 186 bits (473), Expect = 1e-47
Identities = 101/209 (48%), Positives = 138/209 (65%), Gaps = 7/209 (3%)
Frame = +1
Query: 84 SGETV-KRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQL 142
SG T K+KRGRPRKYG D +ALSP P +S P T KRGRG+P S KK
Sbjct: 97 SGSTEGKKKRGRPRKYGPD--CKVALSPMPISASIPLTGDFSAWKRGRGKPLESIKKSFK 270
Query: 143 ------ASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSS 196
A G+ ++ S G F PH++ + GED+ KI++FSQ +A+C+LS++G++S+
Sbjct: 271 FYEAGGAGPGDGIAYSVGANFTPHILTVNEGEDVTMKIMSFSQQGCQAICILSANGTISN 450
Query: 197 VIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVG 256
V +R+P+ SGGTL YEG F I+S+SG Y+ TENG +++R GG+SISL PDGR+ GG +
Sbjct: 451 VTLRQPTSSGGTLTYEGRFEILSLSGSYITTENGLTKSRSGGMSISLAAPDGRVMGGGLA 630
Query: 257 GPLVAASPVQVMIGSFLWGRLKAKNKKKE 285
G LVAA PVQV++ SFL G + K K+
Sbjct: 631 GLLVAAGPVQVVVASFLPGHQLEQQKPKK 717
>TC229041 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, partial (17%)
Length = 787
Score = 182 bits (463), Expect = 1e-46
Identities = 90/134 (67%), Positives = 104/134 (77%)
Frame = +1
Query: 213 GHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF 272
G F I+ +SG Y+ ++G SRNR GGLS+SL PDGR+ GG VGG L+AASPVQV++GSF
Sbjct: 46 GRFEIVCLSGSYLVADSGGSRNRTGGLSVSLASPDGRVVGGGVGGVLIAASPVQVILGSF 225
Query: 273 LWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPTSSLSPWSAASRQM 332
WG K K KKKE SE AE +E+DHQ HNP A+NSISPNQNLTPTSSLSPW ASR +
Sbjct: 226 SWGASKTKIKKKEGSEGAEVALETDHQTVHNPVAVNSISPNQNLTPTSSLSPW-PASRSL 402
Query: 333 DMGNSHADIDLMRG 346
DM NSH DIDLMRG
Sbjct: 403 DMRNSHIDIDLMRG 444
>TC216109 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, partial (35%)
Length = 1141
Score = 178 bits (451), Expect = 4e-45
Identities = 111/265 (41%), Positives = 153/265 (56%), Gaps = 25/265 (9%)
Frame = +3
Query: 104 VSLALSPSPTPSSNPGTMTQGGPKRG--RGRPPGSGKKQQLASFGELMSGSAGTGFIPHV 161
V++ALSP P SS P + KRG RG KK L G+L + S GT F+PH+
Sbjct: 9 VTMALSPMPISSSAPPSNDFSSGKRGKMRGMDYKPSKKVGLDYLGDLNACSDGTNFMPHI 188
Query: 162 IEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMS 221
I + +GEDI K+++FSQ RA+C+LS++G +S+V +R+P SGGTL YEG F I+S+S
Sbjct: 189 ITVNAGEDITMKVISFSQQGPRAICILSANGVISNVTLRQPDSSGGTLTYEGRFEILSLS 368
Query: 222 GCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFL-WGRLKAK 280
G ++PT+N +R+R GG+S+SL PDGR+ GG V G LVAASPVQV++GSFL + + K
Sbjct: 369 GSFMPTDNQGTRSRTGGMSVSLASPDGRVVGGGVAGLLVAASPVQVVVGSFLPSSQQEQK 548
Query: 281 NKKKESSEDAEGTVE--------------------SDHQGAHNPAALNSISPNQNLTPTS 320
KK +SS+ A TV + GAH NS + N NLTP +
Sbjct: 549 IKKPKSSDYAPVTVTPAIAVSSAPPPPTNAEKEDVNVMGGAH--VLQNSGTLNSNLTPPN 722
Query: 321 SL--SPWSAASRQMDMGNSHADIDL 343
+ W D S DI++
Sbjct: 723 AFRRDNWVNMHSMPDSRKSATDINI 797
>TC217941 similar to UP|Q9FIR1 (Q9FIR1) Similarity to AT-hook DNA-binding
protein, partial (48%)
Length = 1479
Score = 174 bits (440), Expect = 7e-44
Identities = 99/230 (43%), Positives = 139/230 (60%), Gaps = 14/230 (6%)
Frame = +1
Query: 89 KRKRGRPRKYGADRVVSLALSPSPT--PSS----------NPGTMTQGGP-KRGRGRPPG 135
K+KRGRPRKY D ++L L+P+ T P+S + GT + P K+ RGRPPG
Sbjct: 364 KKKRGRPRKYSPDGSIALGLAPTHTSPPASAAAGGGSAGDSAGTASADAPAKKHRGRPPG 543
Query: 136 SGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVS 195
SGKKQ A G+ G GF PHVI + SGEDI AKI+ FSQ R +C+LS+ G++
Sbjct: 544 SGKKQLDAL------GAGGVGFTPHVIMVESGEDITAKIMAFSQQGPRTVCILSAIGAIG 705
Query: 196 SVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAV 255
+V +R+P++SGG YEG F I+S+SG +EN R+R L+++L G DGR+ GG V
Sbjct: 706 NVTLRQPAMSGGIATYEGRFEIISLSGSLQQSENNGDRSRTCTLNVTLAGSDGRVLGGGV 885
Query: 256 GGPLVAASPVQVMIGSFL-WGRLKAKNKKKESSEDAEGTVESDHQGAHNP 304
G L AAS VQV++GSF+ + + + + K S A ++ H+P
Sbjct: 886 AGTLTAASTVQVIVGSFIAFAKKSSSSALKSGSSSAPPPXNANLWCTHDP 1035
>TC206704 similar to UP|Q94F52 (Q94F52) At2g33620/F4P9.39, partial (28%)
Length = 1556
Score = 164 bits (414), Expect = 7e-41
Identities = 114/330 (34%), Positives = 169/330 (50%), Gaps = 33/330 (10%)
Frame = +1
Query: 28 QPELHVSPSFNQLSNPNLPFQS-----NIGGGGSNIGTTLPLESSAISSQGVNMSGHTGV 82
QP+LH + + ++ F NI GG S+ L G+ V
Sbjct: 181 QPQLHQNMRMDYAADGTAVFAPPTVTVNINGGDSSPAVPPGL--------GLPQPQPMMV 336
Query: 83 PSGETVKRKRGRPRKYGADRVVSLALSPSPTP-------SSNPGTMTQG--------GPK 127
S E +KRKRGRPRKYG D ++L + TP + G G G
Sbjct: 337 NSPEPIKRKRGRPRKYGPDGGMTLGALKTTTPPGGGVPVGQSGGAFPAGPLSDSASAGTV 516
Query: 128 RGRGRPPGS-GKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALC 186
+ RGRP GS K ++ S G+ F PHVI + +GED++A+I+T SQ +R +C
Sbjct: 517 KRRGRPRGSVNKNKKNDSSNSSKYSGPGSWFTPHVITVNAGEDLSARIMTISQSSSRNIC 696
Query: 187 VLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGP 246
+L+++G++S+V +R+P+ SGGT+ YEG F I+S+ G + + R GGLS+SL GP
Sbjct: 697 ILTANGAISNVTLRQPASSGGTVTYEGRFEILSLGGSFFL----AGTERAGGLSVSLSGP 864
Query: 247 DGRLFGGAVGGPLVAASPVQVMIGSF-------LWGRLKAKNKK-----KESSEDAEGTV 294
DGR+ GG V G L+AASPVQ+++ SF L K +N+K +SS + GT+
Sbjct: 865 DGRVLGGGVAGLLIAASPVQIVLASFVSDVRKHLKRAKKTENEKVSTAGGQSSSPSRGTL 1044
Query: 295 ESDHQGAHNPAALNSISPNQNLTPTSSLSP 324
G + + LN + N T +S +P
Sbjct: 1045SESSGGVGSGSPLNQSTGACNNTIENSTTP 1134
>TC212904 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, partial (35%)
Length = 464
Score = 158 bits (399), Expect = 4e-39
Identities = 74/134 (55%), Positives = 104/134 (77%), Gaps = 1/134 (0%)
Frame = +1
Query: 152 SAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKY 211
S G F PHV+ + +GED+ KI+TFSQ +RA+C+LS++G++S+V +R+PS GGTL Y
Sbjct: 55 SVGANFTPHVLTVNAGEDVTMKIMTFSQQGSRAICILSATGTISNVTLRQPSSCGGTLTY 234
Query: 212 EGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGS 271
EG F I+S+SG ++PTENG +R+R GG+S+SL GPDGR+ GG + G LVAA PVQV++ S
Sbjct: 235 EGRFEILSLSGSFMPTENGVTRSRSGGMSVSLAGPDGRVMGGGLAGLLVAAGPVQVVVAS 414
Query: 272 FLWG-RLKAKNKKK 284
FL G +L+ K KK+
Sbjct: 415 FLPGHQLEHKTKKQ 456
>TC216612 similar to UP|Q8VYJ2 (Q8VYJ2) AT4g12080/F16J13_150, partial (37%)
Length = 906
Score = 157 bits (398), Expect = 5e-39
Identities = 83/206 (40%), Positives = 127/206 (61%), Gaps = 3/206 (1%)
Frame = +3
Query: 141 QLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIR 200
+L + GE S PH+ + SGED+ K+++FSQ RA+C+LS++G +SSV +R
Sbjct: 18 ELENLGEXXXCSXXANXTPHIXXVNSGEDVTMKVISFSQQGPRAICILSANGVISSVTLR 197
Query: 201 EPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLV 260
+P SGGTL YEG F I+S+SG ++P+E+G +R+R GG+S+SL PDGR+ GG V G LV
Sbjct: 198 QPDSSGGTLTYEGRFEILSLSGSFMPSESGGTRSRSGGMSVSLASPDGRVVGGGVAGLLV 377
Query: 261 AASPVQVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAA---LNSISPNQNLT 317
AASPVQV++GSFL G + +K+ E + PAA ++++ P L+
Sbjct: 378 AASPVQVVVGSFLAGNQHEQKPRKQRHEVITSVI---------PAAVVPISTLDPVPILS 530
Query: 318 PTSSLSPWSAASRQMDMGNSHADIDL 343
SS+ + ++ + N ADI++
Sbjct: 531 AASSIRNDNWSAMPAEAKNKPADINV 608
>TC205363 similar to UP|Q9ZRR7 (Q9ZRR7) SAP1 protein, partial (59%)
Length = 1352
Score = 153 bits (386), Expect = 1e-37
Identities = 94/253 (37%), Positives = 139/253 (54%)
Frame = +3
Query: 86 ETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGSGKKQQLASF 145
E KRKRGRPRKYG +LA + T SS + + + K+ L+
Sbjct: 225 EPAKRKRGRPRKYGTPEQ-ALAAKKAATTSSQSFSADKKPHSPTFPSSSFTSSKKSLS-- 395
Query: 146 GELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSIS 205
G+AG GF PHVI +A+GED+ KI+ F Q R +C+LS+SGS+S+ +R+P+ S
Sbjct: 396 --FALGNAGQGFTPHVISVAAGEDVGQKIMLFMQQSRREMCILSASGSISNASLRQPATS 569
Query: 206 GGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPV 265
GG++ YEG F I+S++G YV E G+ R GGLS+ L DG++ GG VGGPL AA PV
Sbjct: 570 GGSIAYEGRFEIISLTGSYVRNELGT---RTGGLSVCLSNTDGQIIGGGVGGPLKAAGPV 740
Query: 266 QVMIGSFLWGRLKAKNKKKESSEDAEGTVESDHQGAHNPAALNSISPNQNLTPTSSLSPW 325
QV++G+F + KK++ +G + + ++S+ Q++ +SS +P
Sbjct: 741 QVIVGTFF------IDNKKDNGAGLKGDASASKLPSPVSEPVSSLGFRQSV-DSSSGNPI 899
Query: 326 SAASRQMDMGNSH 338
M SH
Sbjct: 900 RGNDEHQAMDGSH 938
>AI442988 similar to GP|2213534|emb| DNA-binding PD1-like protein {Pisum
sativum}, partial (8%)
Length = 325
Score = 120 bits (301), Expect = 9e-28
Identities = 68/111 (61%), Positives = 78/111 (70%)
Frame = +1
Query: 26 GAQPELHVSPSFNQLSNPNLPFQSNIGGGGSNIGTTLPLESSAISSQGVNMSGHTGVPSG 85
G QPELH SP+ LSN NLPFQS+IGGGG+ IG+TLPLESS IS+ VN+S +G G
Sbjct: 1 GNQPELHNSPNIRPLSNSNLPFQSSIGGGGT-IGSTLPLESSGISAPCVNVSAPSGAVPG 177
Query: 86 ETVKRKRGRPRKYGADRVVSLALSPSPTPSSNPGTMTQGGPKRGRGRPPGS 136
ETVKRKRGRPRKYG D V S + +PG + Q G KRGRGRPP S
Sbjct: 178 ETVKRKRGRPRKYGPDGAVRCIDSDT---GYHPGALAQ-GQKRGRGRPPWS 318
>TC207820 similar to UP|O04696 (O04696) DNA-binding protein PD1, partial
(38%)
Length = 830
Score = 120 bits (301), Expect = 9e-28
Identities = 65/176 (36%), Positives = 99/176 (55%), Gaps = 1/176 (0%)
Frame = +3
Query: 172 AKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGS 231
AK+L+ SQ R+ LC++S +G+VSSV +R+P+ + ++ +EG F I+ +SG Y+ E+G
Sbjct: 6 AKLLSLSQQRSTTLCIMSGTGTVSSVTLRQPASTNASVTFEGRFQILCLSGSYLVAEDGG 185
Query: 232 SRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSFLWGRLKAKNKKKESSEDAE 291
NR GG+S+SL DG + GG V L+A PVQVM+ SF++G K K+K + +D
Sbjct: 186 PSNRTGGISVSLSSHDGHVIGGGV-AVLIAGGPVQVMLCSFVYGGSKTKSKLGTTIKDES 362
Query: 292 GTVESDHQGAHNPAALNSISPNQNLTPTSSLSPW-SAASRQMDMGNSHADIDLMRG 346
H A P QN P S+ + W + ++ + H IDL RG
Sbjct: 363 SEPPPQHNDKLASPASAPAPPGQNYLPLSATNLWPGSRPAELKSTHMHTGIDLTRG 530
>BG043009 weakly similar to GP|17979485|gb| AT4g12080/F16J13_150 {Arabidopsis
thaliana}, partial (25%)
Length = 310
Score = 119 bits (298), Expect = 2e-27
Identities = 55/92 (59%), Positives = 72/92 (77%)
Frame = +3
Query: 169 DIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTE 228
DI KI+ FSQ ARA+C+LS++G+VS+V +R+PS SGGT+ YEG F I+ +SG Y+ T+
Sbjct: 33 DITTKIMAFSQQGARAVCILSANGAVSTVTLRQPSTSGGTVTYEGRFEIVCLSGSYLVTD 212
Query: 229 NGSSRNRDGGLSISLLGPDGRLFGGAVGGPLV 260
NG SRNR GGLS+SL PDGR+ GG VGG L+
Sbjct: 213 NGGSRNRTGGLSVSLASPDGRVIGGGVGGVLI 308
>TC229102 similar to UP|Q9LTA2 (Q9LTA2) Similarity to AT-hook DNA-binding
protein, partial (63%)
Length = 1141
Score = 104 bits (260), Expect = 5e-23
Identities = 59/168 (35%), Positives = 96/168 (57%), Gaps = 8/168 (4%)
Frame = +1
Query: 113 TPSSNPGTMTQGGP-------KRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIA 165
TPS+ + GG +R RGRPPGS K + +++ P+++E++
Sbjct: 292 TPSTVQKANSSGGDGATIEVVRRPRGRPPGSKNKPKPPV---IITRDPEPAMSPYILEVS 462
Query: 166 SGEDIAAKILTFSQVRARALCVLSSSGSVSSVIIREPSIS-GGTLKYEGHFHIMSMSGCY 224
G D+ I FS+ + +CVL+ SG+V++V +R+PS + G T+ + G F I+S+S +
Sbjct: 463 GGNDVVEAIAQFSRRKNMGICVLTGSGTVANVTLRQPSTTPGSTVTFHGRFDILSVSATF 642
Query: 225 VPTENGSSRNRDGGLSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF 272
+P ++G+S G +ISL GP G++ GG V G L+AA V V+ SF
Sbjct: 643 LPQQSGASPAVPNGFAISLAGPQGQIVGGLVAGGLMAAGTVFVIAASF 786
>TC217688 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like, partial
(61%)
Length = 722
Score = 98.2 bits (243), Expect = 5e-21
Identities = 59/154 (38%), Positives = 86/154 (55%)
Frame = +1
Query: 119 GTMTQGGPKRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFS 178
G + +R RGRPPGS K + F ++ + HV+EIA G DIA + F+
Sbjct: 244 GAIDVATTRRPRGRPPGSRNKPKPPIF---VTRDSPNALRSHVMEIAVGADIADCVAQFA 414
Query: 179 QVRARALCVLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGG 238
+ R R + +LS SG+V +V +R+P+ G + G F I+S++G ++P G S G
Sbjct: 415 RRRQRGVSILSGSGTVVNVNLRQPTAPGAVMALHGRFDILSLTGSFLP---GPSPPGATG 585
Query: 239 LSISLLGPDGRLFGGAVGGPLVAASPVQVMIGSF 272
L+I L G G++ GG V GPL AA PV VM +F
Sbjct: 586 LTIYLAGGQGQIVGGEVVGPLXAAGPVLVMAATF 687
>TC232676 similar to UP|Q6K709 (Q6K709) DNA-binding protein-like, partial
(57%)
Length = 926
Score = 94.0 bits (232), Expect = 9e-20
Identities = 55/146 (37%), Positives = 80/146 (54%)
Frame = +1
Query: 127 KRGRGRPPGSGKKQQLASFGELMSGSAGTGFIPHVIEIASGEDIAAKILTFSQVRARALC 186
+R RGRP GS K S +A HV+EI G D+A I F+ R R +
Sbjct: 19 RRPRGRPAGSKNKXXPPIVITKESPNAXRS---HVLEIXXGSDVAESIAAFANRRHRGVS 189
Query: 187 VLSSSGSVSSVIIREPSISGGTLKYEGHFHIMSMSGCYVPTENGSSRNRDGGLSISLLGP 246
VLS SG V++V +R+P+ G + G F I+S+SG ++P+ + S GL++ L G
Sbjct: 190 VLSGSGIVANVTLRQPAAPAGVITLHGRFEILSLSGAFLPSPSPSGAT---GLTVYLAGG 360
Query: 247 DGRLFGGAVGGPLVAASPVQVMIGSF 272
G++ GG V G LVA+ PV V+ +F
Sbjct: 361 QGQVVGGNVAGSLVASGPVMVIAATF 438
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.130 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,469,933
Number of Sequences: 63676
Number of extensions: 208667
Number of successful extensions: 1560
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 1469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1522
length of query: 346
length of database: 12,639,632
effective HSP length: 98
effective length of query: 248
effective length of database: 6,399,384
effective search space: 1587047232
effective search space used: 1587047232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135311.5


