

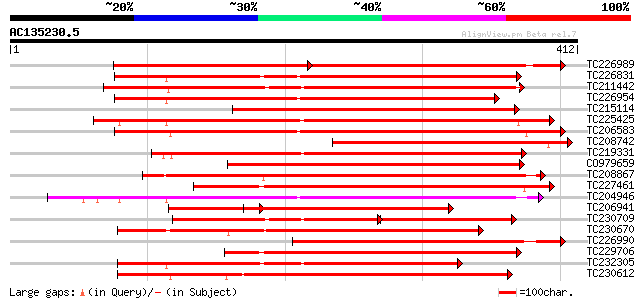
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.5 + phase: 0
(412 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 259 e-129
TC226831 similar to PIR|T52285|T52285 serine/threonine-specific ... 323 9e-89
TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 311 3e-85
TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 310 6e-85
TC215114 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), parti... 305 3e-83
TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T2... 298 2e-81
TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 298 4e-81
TC208742 similar to UP|Q9MAY6 (Q9MAY6) Protein kinase 1, partial... 296 9e-81
TC219331 similar to UP|Q9FM85 (Q9FM85) Protein kinase-like prote... 290 8e-79
CO979659 286 2e-77
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 285 3e-77
TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein ... 283 1e-76
TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B ... 281 5e-76
TC206941 homologue to UP|Q6TKQ5 (Q6TKQ5) Protein kinase-like pro... 275 4e-74
TC230709 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicite... 191 3e-73
TC230670 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 271 5e-73
TC226990 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-prot... 269 1e-72
TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protei... 263 1e-70
TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase... 259 1e-69
TC230612 similar to GB|BAA98102.1|8978074|AB025609 protein serin... 255 3e-68
>TC226989 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (86%)
Length = 1730
Score = 259 bits (661), Expect(2) = e-129
Identities = 125/188 (66%), Positives = 149/188 (78%)
Frame = +3
Query: 217 RDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKS 276
RD K SNILL E YH KLSDFGLAK+GP+GDK+HVSTRVMGTYGYCAP+YAMTGQLT KS
Sbjct: 660 RDCKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKS 839
Query: 277 DIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRG 336
D+YSFGV LELITGRKAID +P EQNLV WARPLF DRR+FS++ DP L+G++P+RG
Sbjct: 840 DVYSFGVVFLELITGRKAIDSTQPQGEQNLVTWARPLFNDRRKFSKLADPRLQGRFPMRG 1019
Query: 337 LYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDPQIHHIQGSRKGSSSPRSRSER 396
LYQALA+A+MC+QE RP+I DVVTAL+YLA+Q YDP G R S R+R ++
Sbjct: 1020LYQALAVASMCIQESAATRPLIGDVVTALSYLANQAYDP-----NGYRGSSDDKRNRDDK 1184
Query: 397 HRRVTSKD 404
R++ D
Sbjct: 1185GGRISKND 1208
Score = 221 bits (562), Expect(2) = e-129
Identities = 104/145 (71%), Positives = 124/145 (84%)
Frame = +2
Query: 76 AQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVV 135
AQTFTF ELAAAT+NFR + FVGEGGFG+VYKG LE Q+VA+KQLD+NG+QG REF+V
Sbjct: 236 AQTFTFRELAAATKNFRPESFVGEGGFGRVYKGRLETTAQIVAVKQLDKNGLQGNREFLV 415
Query: 136 EVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKI 195
EV+ L L HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLHDL P ++PLDWNTRMKI
Sbjct: 416 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEPLDWNTRMKI 595
Query: 196 AAGAARGLEYLHDKMKPPVIYRDLK 220
A GAA+GLEYLHDK PP I + L+
Sbjct: 596 AVGAAKGLEYLHDKANPPGIQQRLQ 670
>TC226831 similar to PIR|T52285|T52285 serine/threonine-specific protein
kinase APK2a [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (82%)
Length = 1693
Score = 323 bits (828), Expect = 9e-89
Identities = 164/305 (53%), Positives = 210/305 (68%), Gaps = 9/305 (2%)
Frame = +1
Query: 77 QTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEK---------INQVVAIKQLDRNGV 127
+ FTF EL AT NFR D +GEGGFG VYKG++++ VVA+K+L G+
Sbjct: 433 KAFTFNELKNATRNFRPDSLLGEGGFGYVYKGWIDEHTFTASKPGSGMVVAVKKLKPEGL 612
Query: 128 QGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPL 187
QG +E++ EV LG H NLVKL+G+CA+GE RLLVYE+M GSLENHL P +PL
Sbjct: 613 QGHKEWLTEVDYLGQLHHQNLVKLIGYCADGENRLLVYEFMSKGSLENHLFRRGP--QPL 786
Query: 188 DWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGD 247
W+ RMK+A GAARGL +LH+ K VIYRD K SNILL ++++KLSDFGLAK GP GD
Sbjct: 787 SWSVRMKVAIGAARGLSFLHNA-KSQVIYRDFKASNILLDAEFNAKLSDFGLAKAGPTGD 963
Query: 248 KTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLV 307
+THVST+VMGT GY AP+Y TG+LT KSD+YSFGV LLEL++GR+A+D K EQNLV
Sbjct: 964 RTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLELLSGRRAVDRSKAGVEQNLV 1143
Query: 308 AWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNY 367
WA+P D+RR ++D L GQYP +G Y A +A C+ + RP I +V+ L
Sbjct: 1144EWAKPYLGDKRRLFRIMDTKLGGQYPQKGAYMAATLALKCLNREAKGRPPITEVLQTLEQ 1323
Query: 368 LASQK 372
+A+ K
Sbjct: 1324IAASK 1338
>TC211442 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (73%)
Length = 1047
Score = 311 bits (797), Expect = 3e-85
Identities = 163/312 (52%), Positives = 213/312 (68%), Gaps = 6/312 (1%)
Frame = +2
Query: 69 VSPDGKVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLE-KIN-----QVVAIKQL 122
+S G FT EL AT +F +GEGGFG VYKG+++ K+ Q VA+K+L
Sbjct: 5 ISFAGSKLYAFTLEELREATNSFSWSNMLGEGGFGPVYKGFVDDKLRSGLKAQTVAVKRL 184
Query: 123 DRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSP 182
D +G+QG RE++ E+I LG HP+LVKL+G+C E E RLL+YEYMP GSLEN L
Sbjct: 185 DLDGLQGHREWLAEIIFLGQLRHPHLVKLIGYCYEDEHRLLMYEYMPRGSLENQLFRRYS 364
Query: 183 GQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKV 242
P W+TRMKIA GAA+GL +LH+ KP VIYRD K SNILL D+ +KLSDFGLAK
Sbjct: 365 AAMP--WSTRMKIALGAAKGLAFLHEADKP-VIYRDFKASNILLDSDFTAKLSDFGLAKD 535
Query: 243 GPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAK 302
GP G+ THV+TR+MGT GY AP+Y MTG LT KSD+YS+GV LLEL+TGR+ +D + +
Sbjct: 536 GPEGEDTHVTTRIMGTQGYAAPEYIMTGHLTTKSDVYSYGVVLLELLTGRRVVDKSRSNE 715
Query: 303 EQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVV 362
++LV WARPL RD+++ +ID LEGQ+P++G + +A C+ PN RP ++DV+
Sbjct: 716 GKSLVEWARPLLRDQKKVYNIIDRRLEGQFPMKGAMKVAMLAFKCLSHHPNARPTMSDVI 895
Query: 363 TALNYLASQKYD 374
L L Q YD
Sbjct: 896 KVLEPL--QDYD 925
>TC226954 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(80%)
Length = 1175
Score = 310 bits (795), Expect = 6e-85
Identities = 155/289 (53%), Positives = 203/289 (69%), Gaps = 9/289 (3%)
Frame = +2
Query: 77 QTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEK---------INQVVAIKQLDRNGV 127
++F++ EL AAT NFR D +GEGGFG V+KG++++ I ++VA+K+L+++G+
Sbjct: 299 KSFSYHELRAATRNFRPDSVLGEGGFGSVFKGWIDEHSLAATKPGIGKIVAVKKLNQDGL 478
Query: 128 QGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPL 187
QG RE++ E+ LG HPNLVKL+G+C E E RLLVYE+MP GS+ENHL +P
Sbjct: 479 QGHREWLAEINYLGQLQHPNLVKLIGYCFEDEHRLLVYEFMPKGSMENHLFRRGSYFQPF 658
Query: 188 DWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGD 247
W+ RMKIA GAA+GL +LH + VIYRD K SNILL +Y++KLSDFGLA+ GP GD
Sbjct: 659 SWSLRMKIALGAAKGLAFLHST-EHKVIYRDFKTSNILLDTNYNAKLSDFGLARDGPTGD 835
Query: 248 KTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLV 307
K+HVSTRVMGT GY AP+Y TG LT KSD+YSFGV LLE+I+GR+AID +P E NLV
Sbjct: 836 KSHVSTRVMGTRGYAAPEYLATGHLTTKSDVYSFGVVLLEMISGRRAIDKNQPTGEHNLV 1015
Query: 308 AWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRP 356
WA+P ++RR ++DP LEGQY A A+A C +P RP
Sbjct: 1016EWAKPYLSNKRRVFRVMDPRLEGQYSQNRAQAAAALAMQCFSVEPKCRP 1162
>TC215114 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (54%)
Length = 878
Score = 305 bits (780), Expect = 3e-83
Identities = 145/208 (69%), Positives = 173/208 (82%)
Frame = +1
Query: 163 LVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCS 222
LVYEYMP+GSLE+HL D P ++PL W+TRMKIA GAARGLEYLH K PPVIYRDLK +
Sbjct: 1 LVYEYMPMGSLEDHLFDPHPDKEPLSWSTRMKIAVGAARGLEYLHCKADPPVIYRDLKSA 180
Query: 223 NILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFG 282
NILL +++ KLSDFGLAK+GP+GD THVSTRVMGTYGYCAP+YAM+G+LT KSDIYSFG
Sbjct: 181 NILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFG 360
Query: 283 VALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALA 342
V LLELITGR+AID + EQNLV+W+R F DR++F +MIDPLL+ +P+R L QA+A
Sbjct: 361 VLLLELITGRRAIDTNRRPGEQNLVSWSRQFFSDRKKFVQMIDPLLQENFPLRCLNQAMA 540
Query: 343 IAAMCVQEQPNMRPVIADVVTALNYLAS 370
I AMC+QEQP RP+I D+V AL YLAS
Sbjct: 541 ITAMCIQEQPKFRPLIGDIVVALEYLAS 624
>TC225425 similar to GB|AAN31120.1|23506217|AY149966 At2g17220/T23A1.8
{Arabidopsis thaliana;} , partial (80%)
Length = 1386
Score = 298 bits (764), Expect = 2e-81
Identities = 158/351 (45%), Positives = 223/351 (63%), Gaps = 16/351 (4%)
Frame = +1
Query: 62 NLNLNDGVSPDGKVAQT-----FTFAELAAATENFRADCFVGEGGFGKVYKGYLEK---- 112
+++ D P+G++ T FTFAEL AAT NF+AD +GEGGFGKV+KG+LE+
Sbjct: 46 SVSSGDQPYPNGQILPTSNLRIFTFAELKAATRNFKADTVLGEGGFGKVFKGWLEEKATS 225
Query: 113 ---INQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMP 169
V+A+K+L+ +QG+ E+ EV LG H NLVKLLG+C E + LLVYE+M
Sbjct: 226 KGGSGTVIAVKKLNSESLQGLEEWQSEVNFLGRLSHTNLVKLLGYCLEESELLLVYEFMQ 405
Query: 170 LGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGED 229
GSLENHL +PL W+ R+KIA GAARGL +LH K VIYRD K SNILL
Sbjct: 406 KGSLENHLFGRGSAVQPLPWDIRLKIAIGAARGLAFLHTSEK--VIYRDFKASNILLDGS 579
Query: 230 YHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELI 289
Y++K+SDFGLAK+GP ++HV+TRVMGT+GY AP+Y TG L KSD+Y FGV L+E++
Sbjct: 580 YNAKISDFGLAKLGPSASQSHVTTRVMGTHGYAAPEYVATGHLYVKSDVYGFGVVLVEIL 759
Query: 290 TGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQ 349
TG +A+D +P+ + L W +P DRR+ ++D LEG++P + ++ ++ C+
Sbjct: 760 TGLRALDSNRPSGQHKLTEWVKPYLHDRRKLKGIMDSRLEGKFPSKAAFRIAQLSMKCLA 939
Query: 350 EQPNMRPVIADVVTALNYL--ASQK-YDPQIHHIQ-GSRKGSSSPRSRSER 396
+P RP + DV+ L + A++K +P+ SR+G + RS R
Sbjct: 940 SEPKHRPSMKDVLENLERIQAANEKPVEPKFRSTHAASRQGHQAVHHRSPR 1092
>TC206583 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(84%)
Length = 1629
Score = 298 bits (762), Expect = 4e-81
Identities = 155/341 (45%), Positives = 219/341 (63%), Gaps = 13/341 (3%)
Frame = +1
Query: 77 QTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQ---------VVAIKQLDRNGV 127
++FT +EL AT NFR D +GEGGFG V+KG++++ + V+A+K+L+++G+
Sbjct: 238 KSFTLSELKTATRNFRPDSVLGEGGFGSVFKGWIDENSLTATKPGTGIVIAVKRLNQDGI 417
Query: 128 QGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPL 187
G RE++ EV LG H +LV+L+GFC E E RLLVYE+MP GS ENHL +PL
Sbjct: 418 PGHREWLAEVNYLGQHFHSHLVRLIGFCLEDEHRLLVYEFMPRGSWENHLFRRGSYFQPL 597
Query: 188 DWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGD 247
W+ R+K+A AA+GL +LH + VIYRD K SN+LL Y++KLSDFGLAK GP GD
Sbjct: 598 SWSLRLKVALDAAKGLAFLHSA-EAKVIYRDFKTSNVLLDSKYNAKLSDFGLAKDGPTGD 774
Query: 248 KTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLV 307
K+HVSTRVMGTYGY AP+Y TG LT KSD+YSFGV LLE+++G++A+D +P+ + NLV
Sbjct: 775 KSHVSTRVMGTYGYAAPEYLATGHLTAKSDVYSFGVVLLEMLSGKRAVDKNRPSGQHNLV 954
Query: 308 AWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNY 367
WA+P ++R+ ++D L+GQY Y+ +A C+ + RP + VVT L
Sbjct: 955 EWAKPFMANKRKIFRVLDTRLQGQYSTDDAYKLATLALRCLSIESKFRPNMDQVVTTLEQ 1134
Query: 368 LASQKYD--PQIHHIQGS-RKGSSSPRS-RSERHRRVTSKD 404
L + P++ +G +P S R RR ++ D
Sbjct: 1135LQLSNVNGGPRVRRRSADVNRGHQNPSSVNGSRVRRRSADD 1257
>TC208742 similar to UP|Q9MAY6 (Q9MAY6) Protein kinase 1, partial (38%)
Length = 691
Score = 296 bits (759), Expect = 9e-81
Identities = 146/177 (82%), Positives = 155/177 (87%), Gaps = 2/177 (1%)
Frame = +2
Query: 235 SDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKA 294
SDFGLAKVGP GDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGV LLELITGRKA
Sbjct: 2 SDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKA 181
Query: 295 IDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNM 354
IDH KPAKEQNL+AWARPLFRDRR+FS M+DPLLEGQYPVRGLYQALAIAAMCVQEQPNM
Sbjct: 182 IDHTKPAKEQNLIAWARPLFRDRRKFSRMVDPLLEGQYPVRGLYQALAIAAMCVQEQPNM 361
Query: 355 RPVIADVVTALNYLASQKYDPQIHHIQGSRKGSSSP--RSRSERHRRVTSKDSETDR 409
RPVI DVVTALNYLASQKYDPQ+H Q SR+ S + + HR + S + ETDR
Sbjct: 362 RPVIVDVVTALNYLASQKYDPQLHPAQTSRRSPPSQIMKRDDDAHRHILSMEHETDR 532
>TC219331 similar to UP|Q9FM85 (Q9FM85) Protein kinase-like protein
(At5g56460), partial (61%)
Length = 1183
Score = 290 bits (742), Expect = 8e-79
Identities = 146/280 (52%), Positives = 198/280 (70%), Gaps = 8/280 (2%)
Frame = +1
Query: 104 KVYKGYL--EKINQV-----VAIKQLDR-NGVQGIREFVVEVITLGLADHPNLVKLLGFC 155
+VYKG++ E I + VA+K D N QG RE++ +V G HPNLVK++G+C
Sbjct: 4 RVYKGFISEELIRKGLPTLDVAVKVHDGDNSHQGHREWLSQVEFWGQLSHPNLVKVIGYC 183
Query: 156 AEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVI 215
E R+L+YEYM G L+N+L +P PL W+ RMKIA GAA+GL +LH+ KP VI
Sbjct: 184 CEDNHRVLIYEYMSRGGLDNYLFKYAPAIPPLSWSMRMKIAFGAAKGLAFLHEAEKP-VI 360
Query: 216 YRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFK 275
YRD K SNILL ++Y+SKLSDFGLAK GP+GDK+HVSTRVMGTYGY AP+Y MTG LT +
Sbjct: 361 YRDFKTSNILLDQEYNSKLSDFGLAKDGPVGDKSHVSTRVMGTYGYAAPEYIMTGHLTPR 540
Query: 276 SDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVR 335
SD+YSFGV LLEL+TGRK++D +PA+EQNL WA PL +++++F +IDP L+G YP++
Sbjct: 541 SDVYSFGVVLLELLTGRKSLDKLRPAREQNLAEWALPLLKEKKKFLNIIDPRLDGDYPIK 720
Query: 336 GLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDP 375
+++A +A C+ P RP++ D+V +L L + P
Sbjct: 721 AVHKAAMLAYHCLNRNPKARPLMRDIVDSLEPLQAHTEVP 840
>CO979659
Length = 731
Score = 286 bits (731), Expect = 2e-77
Identities = 134/216 (62%), Positives = 174/216 (80%)
Frame = -1
Query: 159 EQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRD 218
+ R+LVYE+M GSLENHL D+ ++P+DW RMKIA GAARGLEYLH+ P +IYRD
Sbjct: 731 QHRILVYEFMSNGSLENHLLDIGADKEPMDWKNRMKIAEGAARGLEYLHNGADPAIIYRD 552
Query: 219 LKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDI 278
K SNILL E+++ KLSDFGLAK+GP + HV+TRVMGT+GYCAP+YA +GQL+ KSDI
Sbjct: 551 FKSSNILLDENFNPKLSDFGLAKIGPKEGEEHVATRVMGTFGYCAPEYAASGQLSTKSDI 372
Query: 279 YSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLY 338
YSFGV LLE+ITGR+ D + +EQNL+ WA+PLF+DR +F+ M DPLL+GQ+PV+GL+
Sbjct: 371 YSFGVVLLEIITGRRVFDTARGTEEQNLIDWAQPLFKDRTKFTLMADPLLKGQFPVKGLF 192
Query: 339 QALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYD 374
QALA+AAMC+QE+P+ RP + DVVTAL +LA + +
Sbjct: 191 QALAVAAMCLQEEPDTRPYMDDVVTALAHLAVHRVE 84
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 285 bits (728), Expect = 3e-77
Identities = 152/296 (51%), Positives = 200/296 (67%), Gaps = 3/296 (1%)
Frame = +1
Query: 97 VGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCA 156
+G GGFG VY+G L + VAIK +D+ G QG EF VEV L P L+ LLG+C+
Sbjct: 16 IGHGGFGLVYRGVLND-GRKVAIKFMDQAGKQGEEEFKVEVELLSRLHSPYLLALLGYCS 192
Query: 157 EGEQRLLVYEYMPLGSLENHLHDLSPG---QKPLDWNTRMKIAAGAARGLEYLHDKMKPP 213
+ +LLVYE+M G L+ HL+ +S LDW TR++IA AA+GLEYLH+ + PP
Sbjct: 193 DSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLHEHVSPP 372
Query: 214 VIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLT 273
VI+RD K SNILL + +H+K+SDFGLAK+GP HVSTRV+GT GY AP+YA+TG LT
Sbjct: 373 VIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYALTGHLT 552
Query: 274 FKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYP 333
KSD+YS+GV LLEL+TGR +D K+P E LV+WA PL DR + +++DP LEGQY
Sbjct: 553 TKSDVYSYGVVLLELLTGRVPVDMKRPPGEGVLVSWALPLLTDREKVVKIMDPSLEGQYS 732
Query: 334 VRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDPQIHHIQGSRKGSSS 389
++ + Q AIAAMCVQ + + RP++ADVV +L L + P S+ GSSS
Sbjct: 733 MKEVVQVAAIAAMCVQPEADYRPLMADVVQSLVPLVKTQRSP-------SKVGSSS 879
>TC227461 similar to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (59%)
Length = 1204
Score = 283 bits (723), Expect = 1e-76
Identities = 145/266 (54%), Positives = 188/266 (70%), Gaps = 3/266 (1%)
Frame = +2
Query: 134 VVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRM 193
+ EV LG HP+LVKL+G+C E +QRLLVYE+MP GSLENHL S PL W+ RM
Sbjct: 2 LAEVNYLGDLVHPHLVKLIGYCIEDDQRLLVYEFMPRGSLENHLFRRS---LPLPWSIRM 172
Query: 194 KIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVST 253
KIA GAA+GL +LH++ + PVIYRD K SNILL +Y++KLSDFGLAK GP GDKTHVST
Sbjct: 173 KIALGAAKGLAFLHEEAERPVIYRDFKTSNILLDAEYNAKLSDFGLAKDGPEGDKTHVST 352
Query: 254 RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPL 313
RVMGTYGY AP+Y MTG LT +SD+YSFGV LLE++TGR+++D +P E NLV WARP
Sbjct: 353 RVMGTYGYAAPEYVMTGHLTSRSDVYSFGVVLLEMLTGRRSMDKNRPNGEHNLVEWARPH 532
Query: 314 FRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQK- 372
+RRRF ++IDP LEG + ++G +A +AA C+ P RP++++VV AL L + K
Sbjct: 533 LGERRRFYKLIDPRLEGHFSIKGAQKAAHLAAHCLSRDPKARPLMSEVVEALKPLPNLKD 712
Query: 373 --YDPQIHHIQGSRKGSSSPRSRSER 396
+ + S+SP +R+ R
Sbjct: 713 MASSSYYFQTMQADRFSASPNTRNGR 790
>TC204946 similar to UP|APKB_ARATH (P46573) Protein kinase APK1B , partial
(78%)
Length = 1607
Score = 281 bits (718), Expect = 5e-76
Identities = 153/385 (39%), Positives = 232/385 (59%), Gaps = 24/385 (6%)
Frame = +3
Query: 28 ICFVDKVKVDLNLSELKDKKEDDSK--HDQLSLDVKN-------LNLNDGVSPDGKVAQT 78
+C ++K LN + +D S ++++ DV +++ +G++ Q+
Sbjct: 111 VCLSTQIKAGLNSKHVSADAKDHSSPISNKITKDVSTPISKVSEVSVPQTPRIEGEILQS 290
Query: 79 -----FTFAELAAATENFRADCFVG-EGGFGKVYKGYLEK---------INQVVAIKQLD 123
F+ EL AAT NFR D +G EG FG V+KG+++ VVA+K+L
Sbjct: 291 SNLKNFSLTELTAATRNFRKDSVLGGEGDFGSVFKGWIDNQSLAAAKPGTGVVVAVKRLS 470
Query: 124 RNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPG 183
+ QG ++ + EV LG HP+LVKL+G+C E + RLLVYE+MP GSLENHL
Sbjct: 471 LDSFQGHKDRLAEVNYLGQLSHPHLVKLIGYCFEDKDRLLVYEFMPRGSLENHLFMRGSY 650
Query: 184 QKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVG 243
+PL W R+K+A GAA+GL +LH + VIYRD K SN+LL +Y++KL+D GLAK G
Sbjct: 651 FQPLSWGLRLKVALGAAKGLAFLHSA-ETKVIYRDFKTSNVLLDSNYNAKLADLGLAKDG 827
Query: 244 PIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKE 303
P +K+H STRVMGTYGY AP+Y TG L+ KSD++SFGV LLE+++GR+A+D +P+ +
Sbjct: 828 PTREKSHASTRVMGTYGYAAPEYLATGNLSAKSDVFSFGVVLLEMLSGRRAVDKNRPSGQ 1007
Query: 304 QNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVT 363
NLV WA+P ++R+ ++D LEGQY + + ++ C+ + +RP + +V T
Sbjct: 1008HNLVEWAKPYLSNKRKLLRVLDNRLEGQYELDEACKVATLSLRCLAIESKLRPTMDEVAT 1187
Query: 364 ALNYLASQKYDPQIHHIQGSRKGSS 388
L L Q+ H++ +R+ S+
Sbjct: 1188DLEQL-------QVPHVKQNRRKSA 1241
>TC206941 homologue to UP|Q6TKQ5 (Q6TKQ5) Protein kinase-like protein,
partial (55%)
Length = 852
Score = 275 bits (702), Expect = 4e-74
Identities = 128/152 (84%), Positives = 141/152 (92%)
Frame = +2
Query: 171 GSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDY 230
GSLE+HLHD+SPG+K LDWNTRMKIAAGAARGLEYLHDK PPVIYRDLKCSNILLGE Y
Sbjct: 395 GSLEDHLHDISPGKKQLDWNTRMKIAAGAARGLEYLHDKANPPVIYRDLKCSNILLGEGY 574
Query: 231 HSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELIT 290
H KLSDFGLAK+GP+G+ THVSTRVMGTYGYCAP+YAMTGQLT KSD+YSFGV LLE+IT
Sbjct: 575 HPKLSDFGLAKLGPVGENTHVSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIIT 754
Query: 291 GRKAIDHKKPAKEQNLVAWARPLFRDRRRFSE 322
GRKAID+ K A EQNLVAWARPLF+DRR+FS+
Sbjct: 755 GRKAIDNSKAAGEQNLVAWARPLFKDRRKFSQ 850
Score = 106 bits (264), Expect = 2e-23
Identities = 49/70 (70%), Positives = 61/70 (87%)
Frame = +1
Query: 116 VVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLEN 175
+VAIKQLDRNG+QG REF+VEV+ L L HPNLV L+G+CA+G+QRLLVYE+M LGSLE+
Sbjct: 139 IVAIKQLDRNGLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMSLGSLED 318
Query: 176 HLHDLSPGQK 185
HLH +S G+K
Sbjct: 319 HLHGISIGKK 348
>TC230709 similar to UP|Q84QD9 (Q84QD9) Avr9/Cf-9 rapidly elicited protein
264, partial (61%)
Length = 1245
Score = 191 bits (484), Expect(2) = 3e-73
Identities = 97/153 (63%), Positives = 116/153 (75%)
Frame = +3
Query: 119 IKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLH 178
+K LD +G QG +E++ EV+ LG HP+LVKL+G+C E E RLLVYEY+P GSLEN L
Sbjct: 3 VKLLDLDGSQGHKEWLTEVVFLGQLRHPHLVKLIGYCCEEEHRLLVYEYLPRGSLENQLF 182
Query: 179 DLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFG 238
+ P W+TRMKIAAGAA+GL +LH+ KP VIYRD K SNILL DY++KLSDFG
Sbjct: 183 RIFTASLP--WSTRMKIAAGAAKGLAFLHEAKKP-VIYRDFKASNILLDSDYNAKLSDFG 353
Query: 239 LAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQ 271
LAK GP GD THVSTRVMGT GY AP+Y MTG+
Sbjct: 354 LAKDGPEGDDTHVSTRVMGTQGYAAPEYIMTGK 452
Score = 103 bits (256), Expect(2) = 3e-73
Identities = 49/99 (49%), Positives = 66/99 (66%)
Frame = +2
Query: 270 GQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLE 329
G LT SD+YSFGV LLEL+TGR+++D +P +EQNLV WAR D R+ S ++DP LE
Sbjct: 563 GHLTAMSDVYSFGVVLLELLTGRRSVDKGRPQREQNLVEWARSALNDSRKLSRIMDPRLE 742
Query: 330 GQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYL 368
GQY G +A A+A C+ +P RP+++ VV L L
Sbjct: 743 GQYSEVGARKAAALAYQCLSHRPRSRPLMSTVVNVLEPL 859
>TC230670 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (10%)
Length = 1094
Score = 271 bits (692), Expect = 5e-73
Identities = 141/270 (52%), Positives = 189/270 (69%), Gaps = 4/270 (1%)
Frame = +2
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVI 138
F+F++L +AT F VGEGGFG VY+G L++ + VAIKQL+RNG QG +E++ E+
Sbjct: 287 FSFSDLKSATRAFSRALLVGEGGFGSVYRGLLDQND--VAIKQLNRNGHQGHKEWINELN 460
Query: 139 TLGLADHPNLVKLLGFCAE----GEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMK 194
LG+ HPNLVKL+G+CAE G QRLLVYE+MP SLE+HL P + W TR++
Sbjct: 461 LLGVVKHPNLVKLVGYCAEDDERGIQRLLVYEFMPNKSLEDHLLARVPSTI-IPWGTRLR 637
Query: 195 IAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTR 254
IA AARGL YLH++M +I+RD K SNILL E++++KLSDFGLA+ GP +VST
Sbjct: 638 IAQDAARGLAYLHEEMDFQLIFRDFKTSNILLDENFNAKLSDFGLARQGPSEGSGYVSTA 817
Query: 255 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLF 314
V+GT GY AP+Y TG+LT KSD++SFGV L ELITGR+A+D P EQ L+ W RP
Sbjct: 818 VVGTIGYAAPEYVQTGKLTAKSDVWSFGVVLYELITGRRAVDRNLPRNEQKLLDWVRPYV 997
Query: 315 RDRRRFSEMIDPLLEGQYPVRGLYQALAIA 344
D R+F ++DP L+GQY ++ ++ +A
Sbjct: 998 SDPRKFHHILDPRLKGQYCIKSAHKLAILA 1087
>TC226990 similar to UP|PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase
PBS1 (AvrPphB susceptible protein 1) , partial (50%)
Length = 991
Score = 269 bits (688), Expect = 1e-72
Identities = 130/199 (65%), Positives = 154/199 (77%)
Frame = +1
Query: 206 LHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPD 265
LHDK PPVIYRD K SNILL E YH KLSDFGLAK+GP+GDK+HVSTRVMGTYGYCAP+
Sbjct: 4 LHDKANPPVIYRDFKSSNILLDEGYHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPE 183
Query: 266 YAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMID 325
YAMTGQLT KSD+YSFGV LELITGRKAID +P EQNLV WARPLF DRR+F ++ D
Sbjct: 184 YAMTGQLTVKSDVYSFGVVFLELITGRKAIDSTRPHGEQNLVTWARPLFSDRRKFPKLAD 363
Query: 326 PLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDPQIHHIQGSRK 385
P L+G+YP+RGLYQALA+A+MC+QEQ RP+I DVVTAL++LA+Q YD R
Sbjct: 364 PQLQGRYPMRGLYQALAVASMCIQEQAAARPLIGDVVTALSFLANQAYD--------HRG 519
Query: 386 GSSSPRSRSERHRRVTSKD 404
++R ++ R+ D
Sbjct: 520 AGDDKKNRDDKGGRILKND 576
>TC229706 homologue to UP|Q84SA6 (Q84SA6) Serine/threonine protein kinase,
partial (50%)
Length = 1162
Score = 263 bits (671), Expect = 1e-70
Identities = 130/216 (60%), Positives = 161/216 (74%)
Frame = +2
Query: 157 EGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIY 216
E +QRLLVYE+MP GSLENHL PL W+ RMKIA GAA+GL +LH++ + P+IY
Sbjct: 2 EDDQRLLVYEFMPRGSLENHLFRRP---LPLPWSIRMKIALGAAKGLAFLHEEAQRPIIY 172
Query: 217 RDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKS 276
RD K SNILL +Y++KLSDFGLAK GP G+KTHVSTRVMGTYGY AP+Y MTG LT KS
Sbjct: 173 RDFKTSNILLDAEYNAKLSDFGLAKDGPEGEKTHVSTRVMGTYGYAAPEYVMTGHLTSKS 352
Query: 277 DIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRG 336
D+YSFGV LLE++TGR++ID K+P E NLV WARP+ DRR F +IDP LEG + V+G
Sbjct: 353 DVYSFGVVLLEMLTGRRSIDKKRPNGEHNLVEWARPVLGDRRMFYRIIDPRLEGHFSVKG 532
Query: 337 LYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQK 372
+A +AA C+ P RP++++VV AL L S K
Sbjct: 533 AQKAALLAAQCLSRDPKSRPLMSEVVRALKPLPSLK 640
>TC232305 weakly similar to UP|KPCE_HUMAN (Q02156) Protein kinase C, epsilon
type (nPKC-epsilon) , partial (5%)
Length = 908
Score = 259 bits (663), Expect = 1e-69
Identities = 135/260 (51%), Positives = 175/260 (66%), Gaps = 9/260 (3%)
Frame = +3
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEK---------INQVVAIKQLDRNGVQG 129
+T EL +AT NFR D +GEGGFG+V+KG+++K + VA+K+ + + +QG
Sbjct: 138 YTLDELRSATRNFRPDTVLGEGGFGRVFKGWIDKNTFKPSRVGVGIPVAVKKSNPDSLQG 317
Query: 130 IREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDW 189
+ E+ EV LG HPNL KL+G+C E Q LLVYEYM GSLE+HL P KPL W
Sbjct: 318 LEEWQSEVQLLGKFSHPNLGKLIGYCWEESQFLLVYEYMQKGSLESHLFRRGP--KPLSW 491
Query: 190 NTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKT 249
+ R+KIA GAARGL +LH K VIYRD K SNILL D+++KLSDFGLAK GP+ K+
Sbjct: 492 DIRLKIAIGAARGLAFLHTSEKS-VIYRDFKSSNILLDGDFNAKLSDFGLAKFGPVNGKS 668
Query: 250 HVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAW 309
HV+TR+MGTYGY AP+Y TG L KSD+Y FGV LLE++TGR A+D +P QNLV
Sbjct: 669 HVTTRIMGTYGYAAPEYMATGHLYIKSDVYGFGVVLLEMLTGRAALDTNQPTGMQNLVEC 848
Query: 310 ARPLFRDRRRFSEMIDPLLE 329
++R E++DP +E
Sbjct: 849 TMSSLHAKKRLKEVMDPNME 908
>TC230612 similar to GB|BAA98102.1|8978074|AB025609 protein serine/threonine
kinase-like {Arabidopsis thaliana;} , partial (70%)
Length = 1606
Score = 255 bits (651), Expect = 3e-68
Identities = 134/297 (45%), Positives = 190/297 (63%), Gaps = 10/297 (3%)
Frame = +3
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQ------VVAIKQLDRNGVQGIRE 132
FT EL AT F +GEGGFGKVY+G ++ + +VAIK+L+ G+QG +E
Sbjct: 213 FTLQELVDATHGFNRMLKIGEGGFGKVYRGTIKPHPEDGADPILVAIKKLNTRGLQGHKE 392
Query: 133 FVVEVITLGLADHPNLVKLLGFCA----EGEQRLLVYEYMPLGSLENHLHDLSPGQKPLD 188
++ EV L + +HPNLVKLLG+C+ +G QRLLVYE+M + S L
Sbjct: 393 WLAEVQFLSVVNHPNLVKLLGYCSVDSEKGIQRLLVYEFMS-NRESGKITFFSLSLPHLT 569
Query: 189 WNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDK 248
W TR++I GAA+GL YLH+ ++ VIYRD K SN+LL + +H KLSDFGLA+ GP GD+
Sbjct: 570 WKTRLQIMLGAAQGLHYLHNGLEVKVIYRDFKSSNVLLDKKFHPKLSDFGLAREGPTGDQ 749
Query: 249 THVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVA 308
THVST V+GT GY AP+Y TG L +SDI+SFGV L E++TGR+ ++ +P EQ L+
Sbjct: 750 THVSTAVVGTQGYAAPEYVETGHLKIQSDIWSFGVVLYEILTGRRVLNRNRPIGEQKLIE 929
Query: 309 WARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
W + + RFS++IDP L+ QY + + +A C+++ P RP ++ +V +L
Sbjct: 930 WVKNYPANSSRFSKIIDPRLKNQYSLGAARKVAKLADNCLKKNPEDRPSMSQIVESL 1100
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,790,804
Number of Sequences: 63676
Number of extensions: 217844
Number of successful extensions: 2834
Number of sequences better than 10.0: 965
Number of HSP's better than 10.0 without gapping: 2040
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2071
length of query: 412
length of database: 12,639,632
effective HSP length: 100
effective length of query: 312
effective length of database: 6,272,032
effective search space: 1956873984
effective search space used: 1956873984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135230.5


