

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
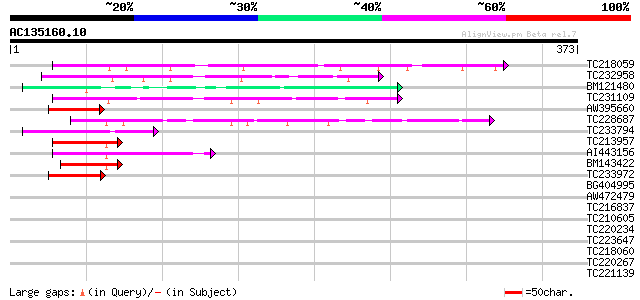
Query= AC135160.10 + phase: 0
(373 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 80 2e-15
TC232958 75 5e-14
BM121480 62 3e-10
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 60 2e-09
AW395660 49 5e-06
TC228687 similar to UP|Q9FGY4 (Q9FGY4) Gb|AAF30317.1, partial (79%) 48 8e-06
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 47 1e-05
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 45 7e-05
AI443156 43 3e-04
BM143422 43 3e-04
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 41 8e-04
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 39 0.003
AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-bind... 39 0.004
TC216837 similar to UP|Q9SGY4 (Q9SGY4) F20B24.9, partial (6%) 33 0.16
TC210605 weakly similar to UP|Q9FV01 (Q9FV01) SKP1 interacting p... 33 0.16
TC220234 similar to GB|AAM65311.1|21593362|AY087775 microtubule-... 32 0.60
TC223647 31 1.0
TC218060 31 1.0
TC220267 similar to UP|Q6K235 (Q6K235) F-box-like protein, parti... 31 1.0
TC221139 31 1.0
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 79.7 bits (195), Expect = 2e-15
Identities = 85/353 (24%), Positives = 146/353 (41%), Gaps = 53/353 (15%)
Frame = +3
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFAT-----------RRNRLLILQNA-- 75
EL +L RLP+K L KCV KSW ++I P+F + + LL+++
Sbjct: 75 ELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHLLVIRRPFF 254
Query: 76 ----PNMKFIFCDGGNDQKSIPIKSLFPQDVAR------IEIYGSCDGVFCLKGISSCIT 125
+ + + + +K + L P EI G C+G++ L+G +
Sbjct: 255 SGLKTYISVLSWNTNDPKKHVSSDVLNPPYEYNSDHKYWTEILGPCNGIYFLEGNPN--- 425
Query: 126 RHDQLILWNPTTKEVHLIPRAPSLGNH----YSDESLYGFGAVNDDFKVVKL-NISNSNR 180
+L NP+ E +P++ H ++D + +GF +D+KVV L ++
Sbjct: 426 -----VLMNPSLGEFKALPKSHFTSPHGTYTFTDYAGFGFDPKTNDYKVVVLKDLWLKET 590
Query: 181 MAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQ---PSRYNTLVNGVYYWITSSDGS 237
+ A++Y L++ SW L P + I+ SR T N +W + S
Sbjct: 591 DEREIGYWSAELYSLNSNSWRKL---DPSLLPLPIEIWGSSRVFTYANNCCHWWGFVEES 761
Query: 238 DAAR--ILCFDFRDNQFRKLEAPKL--------GHYIPFFCDDVFEIKGYLGYVV-QYRC 286
DA + +L FD FRK+ PK+ G +P FE +G++V R
Sbjct: 762 DATQDVVLAFDMVKESFRKIRVPKIRDSSDEKFGTLVP------FEESASIGFLVYPVRG 923
Query: 287 RIVWLEIWTL----EQNGWAKKYNIDTKMSIFHIYG-------LWNDGAEILV 328
++W + ++ W K+Y++ I+ + G W D E LV
Sbjct: 924 TEKRFDVWVMKDYWDEGSWVKQYSVGPVQVIYKLVGFYGTNRFFWKDSNERLV 1082
>TC232958
Length = 758
Score = 75.1 bits (183), Expect = 5e-14
Identities = 73/247 (29%), Positives = 106/247 (42%), Gaps = 22/247 (8%)
Frame = +1
Query: 22 ERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRR---------NRLLIL 72
++M L ++L EILLRLP KSL RFK V KSW +I P FA L I
Sbjct: 49 KKMVLPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIA 228
Query: 73 QNAPNMKFIFCDGG--NDQKSIPIKSLFPQDVAR---IEIYGSCDGVFCLKGISSCITRH 127
+AP ++ I + +D S+ + P +EI GSC G L +S
Sbjct: 229 SSAPELRSIDFNASLHDDSASVAVTVDLPAPKPYFHFVEIIGSCRGFILLHCLS------ 390
Query: 128 DQLILWNPTTKEVHLIPRAPSLGN----HYSDESLYGFGAVNDDFKVVKLNISNSNRMAK 183
L +WNPTT ++P +P N ++ +G+ DDF VV N A
Sbjct: 391 -HLCVWNPTTGVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDDFLVVHA-CYNPKHQAN 564
Query: 184 INSLLKADIYDLSTKSWTPLVS-HPPITMVTRIQPSRYN---TLVNGVYYWITSSDGSDA 239
A+I+ L +W + H P T +RYN + +NG +W+ +
Sbjct: 565 C-----AEIFSLRANAWKGIEGIHFPYTHFR--YTNRYNQFGSFLNGAIHWLAFRINASI 723
Query: 240 ARILCFD 246
I+ FD
Sbjct: 724 NVIVAFD 744
>BM121480
Length = 917
Score = 62.4 bits (150), Expect = 3e-10
Identities = 71/254 (27%), Positives = 103/254 (39%), Gaps = 4/254 (1%)
Frame = +3
Query: 9 KKFSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCV--QKSWNNIIKSPYFATRR 66
K+ R K K + L +EL EILLRLP KSL RFKCV + +++ P
Sbjct: 15 KERKRSMKKKQNQSLTTLPQELIREILLRLPVKSLLRFKCVLFKTCSRDVVYFP------ 176
Query: 67 NRLLILQNAPNMKFIFCDGGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITR 126
L L + P ++ D I R +I GSC G+ L S
Sbjct: 177 ---LPLPSIPCLRL-------DDFGI-----------RPKILGSCRGLVLLYYDDSA--- 284
Query: 127 HDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNRMAKINS 186
LILWNP+ + R L N+ D + + +G D+ K L I + K
Sbjct: 285 --NLILWNPS------LGRHKXLPNYRDDITSFPYGFGYDESKDEYLLILIG--LPKFGP 434
Query: 187 LLKADIYDLSTKSW-TPLVSHPPITMVTRIQP-SRYNTLVNGVYYWITSSDGSDAARILC 244
ADIY T+SW T + + P+ +R +L+NG +W S+ I+
Sbjct: 435 ETGADIYSFKTESWKTDTIVYDPLXRYXAEDXIARAGSLLNGALHWFVFSESKXDHVIIA 614
Query: 245 FDFRDNQFRKLEAP 258
FD + + P
Sbjct: 615 FDLVERTLSXIPLP 656
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 59.7 bits (143), Expect = 2e-09
Identities = 55/240 (22%), Positives = 104/240 (42%), Gaps = 10/240 (4%)
Frame = +3
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFA----TRRNRLLILQNAPNMKFIFCD 84
E+ EIL RLP KS+ R + K W +II S +F + + LIL++ ++ +
Sbjct: 102 EVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKSHSSLILRHRSHLYSLDLK 281
Query: 85 GGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEVHLIP 144
+Q + + I++ GS +G+ C+ ++ D + LWNP ++ ++P
Sbjct: 282 -SPEQNPVELSHPLMCYSNSIKVLGSSNGLLCISNVA------DDIALWNPFLRKHRILP 440
Query: 145 --RAPSLGNHYSDESLYGFG--AVNDDFKVVKLNISNSNRMAKINSLLKADIYDLSTKSW 200
R + +YGFG + ++D+K++ + + +S + +Y L + SW
Sbjct: 441 ADRFHRPQSSLFAARVYGFGHHSPSNDYKLLSITYFVDLQKRTFDS--QVQLYTLKSDSW 614
Query: 201 TPLVSHPPITMVTRIQPSRYNTLVNGVYYWITSS--DGSDAARILCFDFRDNQFRKLEAP 258
L S P R V+G +W+ + + I+ FD F ++ P
Sbjct: 615 KNLPSMPYALCCARTM----GVFVSGSLHWLVTRKLQPHEPDLIVSFDLTRETFHEVPLP 782
>AW395660
Length = 381
Score = 48.5 bits (114), Expect = 5e-06
Identities = 22/37 (59%), Positives = 28/37 (75%)
Frame = +3
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF 62
L +EL +EIL RLP KSL +F+CV KSW ++I PYF
Sbjct: 264 LPDELVVEILSRLPVKSLLQFRCVCKSWMSLIYDPYF 374
>TC228687 similar to UP|Q9FGY4 (Q9FGY4) Gb|AAF30317.1, partial (79%)
Length = 1228
Score = 47.8 bits (112), Expect = 8e-06
Identities = 65/295 (22%), Positives = 123/295 (41%), Gaps = 16/295 (5%)
Frame = +1
Query: 41 KSLSRFKCVQKSWNNIIKSPYF------ATRRNRLLILQ--NAPNMKFIFCDGGNDQKSI 92
KSL RFK V K W + YF +R+N +++++ ++ K N +
Sbjct: 4 KSLFRFKTVCKLWYRLSLDKYFVQLYNEVSRKNPMILVEISDSSESKTSLICVDNLRGVS 183
Query: 93 PIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILWNPTTKEVHLIP--RAPSLG 150
+F D R+++ SC+G+ C S I + NP T+E L+P R +
Sbjct: 184 EFSFIFLND--RVKVRASCNGLLC----CSSIPDKGVFYVCNPVTREYRLLPKSRERHVT 345
Query: 151 NHYSD--ESLYGFGAVNDDFKVVKLNISNSNRM--AKINSLLKADIYDLSTKSWTPLVSH 206
Y D +L G A + ++ + ++ +RM + + ++D W VS
Sbjct: 346 RFYPDGEATLVGL-ACDSAYRKFNVVLAGYHRMFGHRPDGSFICLVFDSELNKWRKFVSF 522
Query: 207 PP--ITMVTRIQPSRYNTLVNGVYYWITSSDGSDAARILCFDFRDNQFRKLEAPKLGHYI 264
T + + Q VN +W+T+S + IL D +RK++ P + +
Sbjct: 523 QDDHFTHMNKNQV----VFVNNALHWLTAS----STYILVLDLSCEVWRKMQLP---YDL 669
Query: 265 PFFCDDVFEIKGYLGYVVQYRCRIVWLEIWTLEQNGWAKKYNIDTKMSIFHIYGL 319
+ + + G + + W+ IW L ++ W ++ + K+S+ I G+
Sbjct: 670 ICGTGNRIYLLDFDGCLSVIKISEAWMNIWVL-KDYWKDEWCMVDKVSLRCIRGM 831
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 47.0 bits (110), Expect = 1e-05
Identities = 34/90 (37%), Positives = 45/90 (49%)
Frame = +1
Query: 9 KKFSRLRKTKLSIERMKLGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNR 68
K+ R K K + L +EL EILLRLP KSL RFKCV KS+ ++I P F
Sbjct: 58 KERKRSMKKKQNQSLTTLPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISH-- 231
Query: 69 LLILQNAPNMKFIFCDGGNDQKSIPIKSLF 98
L +P + I +SI +S+F
Sbjct: 232 -YALAASPTHRLILRSHDFYAQSIATESVF 318
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 44.7 bits (104), Expect = 7e-05
Identities = 24/52 (46%), Positives = 33/52 (63%), Gaps = 6/52 (11%)
Frame = +3
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF------ATRRNRLLILQN 74
EL EILLR P +S+ RFKCV KSW ++I P F A+ +RL+++ N
Sbjct: 42 ELIREILLRSPVRSVLRFKCVCKSWLSLISDPQFTHFDLAASPTHRLILISN 197
>AI443156
Length = 414
Score = 42.7 bits (99), Expect = 3e-04
Identities = 30/111 (27%), Positives = 49/111 (44%), Gaps = 4/111 (3%)
Frame = +3
Query: 29 ELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYF----ATRRNRLLILQNAPNMKFIFCD 84
E+ EIL RLP KS+ R + K W +II S +F + + LIL++ + +
Sbjct: 99 EVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFHLNKSHTSLILRHRSQLYSLDLK 278
Query: 85 GGNDQKSIPIKSLFPQDVARIEIYGSCDGVFCLKGISSCITRHDQLILWNP 135
D + I++ GS +G+ C+ + D + LWNP
Sbjct: 279 SLLDPNPFELSHPLMCYSNSIKVLGSSNGLLCISNVX------DDIALWNP 413
>BM143422
Length = 424
Score = 42.7 bits (99), Expect = 3e-04
Identities = 23/50 (46%), Positives = 30/50 (60%), Gaps = 9/50 (18%)
Frame = +1
Query: 34 ILLRLPTKSLSRFKCVQKSWNNIIKSPYF---------ATRRNRLLILQN 74
ILL+LP KS++RFKCV KSW ++I P F A +RLL+ N
Sbjct: 253 ILLKLPVKSVTRFKCVCKSWLSLISDPQFGFSHFDLALAVPSHRLLLRSN 402
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 41.2 bits (95), Expect = 8e-04
Identities = 22/38 (57%), Positives = 28/38 (72%)
Frame = -2
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFA 63
L +E+ IEILLRLP KSL FK V KS ++I +P+FA
Sbjct: 540 LPQEIIIEILLRLPVKSLPSFKFVCKS*LSLISNPHFA 427
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 39.3 bits (90), Expect = 0.003
Identities = 31/109 (28%), Positives = 51/109 (46%), Gaps = 10/109 (9%)
Frame = +1
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFA-TRRNRLLILQNAPNMKFIFCD 84
L E+ EIL RLP +SL RF+ KSW ++I S + R L L + ++ + D
Sbjct: 40 LPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLASNTSL-ILRVD 216
Query: 85 GGNDQKSIPIKSLFPQDVAR---------IEIYGSCDGVFCLKGISSCI 124
Q + P +L P I + GSC+G+ C+ +++ +
Sbjct: 217 SDLYQTNFP--TLDPPVSLNHPLMCYSNSITLLGSCNGLLCISNVANTV 357
>AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-binding protein
GRP1 - Commerson's wild potato, partial (22%)
Length = 414
Score = 38.9 bits (89), Expect = 0.004
Identities = 22/99 (22%), Positives = 47/99 (47%)
Frame = +2
Query: 111 CDGVFCLKGISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKV 170
C+G+ C+ C+ +I+ NP+ + +P +Y+ GF + N D+KV
Sbjct: 131 CNGLICIAYGERCLP----IIICNPSIRRYVWLPTPHDYPGYYNSCIALGFDSTNCDYKV 298
Query: 171 VKLNISNSNRMAKINSLLKADIYDLSTKSWTPLVSHPPI 209
++++ + +++ L ++Y L + SW L P+
Sbjct: 299 IRISCIVDDESFGLSAPL-VELYSLKSGSWRILDGIAPV 412
>TC216837 similar to UP|Q9SGY4 (Q9SGY4) F20B24.9, partial (6%)
Length = 415
Score = 33.5 bits (75), Expect = 0.16
Identities = 30/110 (27%), Positives = 49/110 (44%)
Frame = +2
Query: 111 CDGVFCLKGISSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKV 170
C+ C IS T + I+W+P L PS+ N +++ N D+
Sbjct: 104 CNLSLCNSFISLSSTITNNKIMWSP------LAYAYPSMTNTFNN---------NKDYNF 238
Query: 171 VKLNISNSNRMAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRY 220
+K+N++N+N M +N LL Y K+W L S+ + +T RY
Sbjct: 239 IKINLNNNNNMG-LNLLLS---YLYYPKAWV-LKSNNSVMRLTNYSMPRY 373
>TC210605 weakly similar to UP|Q9FV01 (Q9FV01) SKP1 interacting partner 4,
partial (12%)
Length = 661
Score = 33.5 bits (75), Expect = 0.16
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 1/46 (2%)
Frame = +1
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKS-PYFATRRNRLL 70
L +++ + L R+P K S KCV K W N+I S +F RR L
Sbjct: 211 LPDDISLMCLARIPRKYHSVMKCVSKRWRNLICSEEWFCYRRKHKL 348
>TC220234 similar to GB|AAM65311.1|21593362|AY087775 microtubule-associated
protein EB1-like protein {Arabidopsis thaliana;} ,
partial (36%)
Length = 917
Score = 31.6 bits (70), Expect = 0.60
Identities = 30/106 (28%), Positives = 45/106 (42%)
Frame = +2
Query: 121 SSCITRHDQLILWNPTTKEVHLIPRAPSLGNHYSDESLYGFGAVNDDFKVVKLNISNSNR 180
SS +R +++ NPT + + APS GN +E K+ +L +S +
Sbjct: 203 SSHNSRRNEVSNANPTNQAAKVARAAPSAGNPAYEE------------KITELKLS-IDS 343
Query: 181 MAKINSLLKADIYDLSTKSWTPLVSHPPITMVTRIQPSRYNTLVNG 226
+ K A + D+ TP + H PI V IQ Y T NG
Sbjct: 344 LEKERDFYFAKLRDIEILCQTPEIEHSPI--VAAIQKILYATDDNG 475
>TC223647
Length = 448
Score = 30.8 bits (68), Expect = 1.0
Identities = 14/31 (45%), Positives = 18/31 (57%)
Frame = +3
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNI 56
L E+L IL RLP + CV KSWN++
Sbjct: 123 LNEDLLQNILARLPALHFASAACVSKSWNSL 215
>TC218060
Length = 901
Score = 30.8 bits (68), Expect = 1.0
Identities = 31/123 (25%), Positives = 46/123 (37%), Gaps = 16/123 (13%)
Frame = +2
Query: 222 TLVNGVYYW--ITSSDGSDAARILCFDFRDNQFRKLEAPKL--GHYIPFFCDDVFEIKGY 277
T N +W G+ +L FD FRK+ PK+ F FE
Sbjct: 8 TYANNCCHWWGFVEDXGATQDIVLAFDMVKESFRKIRVPKVRDSSDEKFATLVPFEESAS 187
Query: 278 LGYVV-QYRCRIVWLEIWTL----EQNGWAKKYN---IDTKMSIFHIYG----LWNDGAE 325
+G +V R ++W + ++ W K+Y+ + I YG LW D E
Sbjct: 188 IGVLVYPVRGAEKSFDVWVMKDYWDEGSWVKQYSVGPVQVNHRIVGFYGTNRFLWKDSNE 367
Query: 326 ILV 328
LV
Sbjct: 368 RLV 376
>TC220267 similar to UP|Q6K235 (Q6K235) F-box-like protein, partial (7%)
Length = 586
Score = 30.8 bits (68), Expect = 1.0
Identities = 16/45 (35%), Positives = 23/45 (50%)
Frame = +2
Query: 26 LGEELEIEILLRLPTKSLSRFKCVQKSWNNIIKSPYFATRRNRLL 70
L ++L IL LP S+ R CV K W+ I+ S F + +L
Sbjct: 338 LPDDLLERILAYLPIASIFRAGCVSKRWHEIVNSERFVWNLSHVL 472
>TC221139
Length = 634
Score = 30.8 bits (68), Expect = 1.0
Identities = 14/30 (46%), Positives = 19/30 (62%)
Frame = +3
Query: 33 EILLRLPTKSLSRFKCVQKSWNNIIKSPYF 62
EILLR+P ++S+ V K W +I SP F
Sbjct: 129 EILLRIPPPTISKLIIVSKIWLRVICSPSF 218
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.140 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,399,877
Number of Sequences: 63676
Number of extensions: 323841
Number of successful extensions: 1532
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1520
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1527
length of query: 373
length of database: 12,639,632
effective HSP length: 99
effective length of query: 274
effective length of database: 6,335,708
effective search space: 1735983992
effective search space used: 1735983992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC135160.10


