

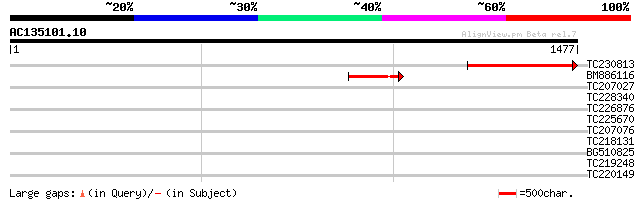
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135101.10 + phase: 0 /pseudo
(1477 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC230813 454 e-127
BM886116 185 1e-46
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 33 0.93
TC228340 similar to UP|Q9CJT2 (Q9CJT2) OppB, partial (5%) 33 0.93
TC226876 similar to UP|Q84L33 (Q84L33) RAD23-like protein, parti... 32 2.7
TC225670 UP|P93167 (P93167) Asparagine synthetase 2 , complete 31 3.5
TC207076 similar to UP|Q8LAJ0 (Q8LAJ0) MutT domain protein-like,... 30 7.9
TC218131 similar to UP|Q9LFZ9 (Q9LFZ9) F20N2.11, partial (8%) 30 7.9
BG510825 30 7.9
TC219248 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich... 30 7.9
TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferas... 30 7.9
>TC230813
Length = 1130
Score = 454 bits (1169), Expect = e-127
Identities = 225/284 (79%), Positives = 252/284 (88%)
Frame = +1
Query: 1194 PSPIQRVSLTWKLHSLSVNFLVGMEILEQDQGRETFEALQDLYGELLDKERFNQNKEAIS 1253
PSP+Q+VSLTWKLHSLSVNFLVGMEILEQD R+ FEALQDLYGELLD R NQ+K IS
Sbjct: 1 PSPVQQVSLTWKLHSLSVNFLVGMEILEQDWSRDIFEALQDLYGELLDNARLNQSKYFIS 180
Query: 1254 DDKKHIEFLRFKSDIHESYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCCVESSIRLAT 1313
DDKKH+EFLRF+++IHESYSTF+EELVEQFS++SYGD+IFGRQVS+YLH CVE+SIRLA
Sbjct: 181 DDKKHLEFLRFQTEIHESYSTFLEELVEQFSAVSYGDVIFGRQVSLYLHRCVETSIRLAA 360
Query: 1314 WNTLSNARVLELLPPLEKCFSGAEGYLEPAEDNEEILEAYAKSWVSDALDRAEIRGSVSY 1373
WNTLSN+RVLELLPPLEKCFSGAEGYLEPAEDNE ILEAY WVSDALDRA IRGSV+Y
Sbjct: 361 WNTLSNSRVLELLPPLEKCFSGAEGYLEPAEDNEAILEAYTNLWVSDALDRAAIRGSVAY 540
Query: 1374 TMAVHHLSSFIFNACPVDKLLLRNNLVRSLLRDYAGKQQHEGMLMNLISHNRQSTSNMDE 1433
T+ VHHLSSFIF+ACP DKLLLRN L RSLLRDYAGKQQHEGML+NLI HN+ S M E
Sbjct: 541 TLVVHHLSSFIFHACPTDKLLLRNRLARSLLRDYAGKQQHEGMLLNLIHHNKPPPSVMGE 720
Query: 1434 QLDGLLHEESWLESRMKVLIEACEGNSSLLIQVKKLKDAAEKNS 1477
+L+G+L E+SWLESR+KVL+EACEGNSS+L V KLK A KNS
Sbjct: 721 ELNGILSEKSWLESRLKVLVEACEGNSSILTVVDKLK-AVVKNS 849
>BM886116
Length = 432
Score = 185 bits (470), Expect = 1e-46
Identities = 93/143 (65%), Positives = 108/143 (75%)
Frame = +3
Query: 884 GICNPPVTEQSLSKEGKVLEEGIVSRCLVELRSMLDVFTFSASSGWQRMQSIEIFGRGGP 943
GIC+ P EQSLSKEGKVLE+GIV+ CLVELR MLD F FS SSGW +QSIE FGRGGP
Sbjct: 6 GICSLPRQEQSLSKEGKVLEDGIVNGCLVELRYMLDAFMFSVSSGWHHIQSIESFGRGGP 185
Query: 944 APGMGVGWGAHGGGFWSKTVLPVKTDARLLVCLLQIFENTSNDAPETEQMTFSMQQVNTA 1003
PG+ +GWGA GGFWS T L + DA+ LV LL+IFEN S TE+ TF +Q+VN
Sbjct: 186 VPGV*IGWGAPSGGFWSATFLLAQIDAKFLVSLLEIFENASKGV-VTEETTFIIQRVNAG 362
Query: 1004 LGLCLTAGPADMVVIEKTLDLLF 1026
LGLCLTAGP + VV++K LDLLF
Sbjct: 363 LGLCLTAGPREKVVVQKALDLLF 431
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 33.1 bits (74), Expect = 0.93
Identities = 58/297 (19%), Positives = 111/297 (36%), Gaps = 18/297 (6%)
Frame = +3
Query: 25 KWREKKTKGMDFGKWREFTQDDKSFLGKDLE-KDVSSYGPTTGRKKNENGGKNTSKKISS 83
K ++KK K K + D + GK+ + +D ++K + GK ++KI
Sbjct: 24 KEKDKKEKKKKENKDKGDKTDVEKVKGKENDGEDDEEKKEKKKKEKKDKDGKTDAEKIKK 203
Query: 84 YSDGSVFASMEVDAKPQLVKLDGGFINSATSMELDTSNKDDKKEVFAAERDKIFSDRMTD 143
D + + K + K+D + D NK +KK+ + D D
Sbjct: 204 KEDDGKDDEEKKEKKKKYDKIDAXKVKGKEDDGKDEGNK-EKKDKEKGDGDGEEKKEKKD 380
Query: 144 HSSTSEKNYFMHEQESTSL-------ENEIDSENRARIQQMSTEEIEEAKADIMEKISPA 196
+K ++E+ +L E E D N+ + + +E + K
Sbjct: 381 KEKEKKKEKKDKDEETDTLKEKGKNDEGEDDEGNKKKKKDKKEKEKDHKK---------- 530
Query: 197 LLKVLQKRGKEKLKKPNSLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHTIMAPPSKK 256
+K+ KE+ +K +S K EV + + I + K + ED KK
Sbjct: 531 -----EKKDKEEGEKEDS-KVEVSV------RDIDIEEIKKEGEKEDKGKDGGKEVKEKK 674
Query: 257 QLDDKN-------VSGKTST---TTSSSSWNAWSNRVEAIRELRFSLAGDVVDTEQE 303
+ +DK+ V+GK T +T + +++ + E + + + + E E
Sbjct: 675 KKEDKDKKEKKKKVTGKDKTKDLSTLKQKLEKINGKIQPLLEKKADIERQIKEVEAE 845
>TC228340 similar to UP|Q9CJT2 (Q9CJT2) OppB, partial (5%)
Length = 466
Score = 33.1 bits (74), Expect = 0.93
Identities = 27/112 (24%), Positives = 47/112 (41%), Gaps = 4/112 (3%)
Frame = +2
Query: 15 RKKTKGMDFGKWREKKTKGMDFGKWREFTQDDKSFLGKDLEKDVSSYGPTTGRKKNENGG 74
+K TK + K ++KKTK + K + + + +K ++ P T +KKN
Sbjct: 110 KKITKNTEISKTQKKKTKNAETPKTEKKKTKNAETPKTEKKKTKNAETPKTEKKKNAEIP 289
Query: 75 KNTSKKISS----YSDGSVFASMEVDAKPQLVKLDGGFINSATSMELDTSNK 122
K KK S +DG E+ + K +++ TS + D +K
Sbjct: 290 KIEKKKTKSAEIPLTDGKTKRDREIAKEENKKKTKKPKLSNGTSKQRDEGSK 445
>TC226876 similar to UP|Q84L33 (Q84L33) RAD23-like protein, partial (87%)
Length = 1499
Score = 31.6 bits (70), Expect = 2.7
Identities = 18/69 (26%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Frame = +2
Query: 190 MEKISPALLK-VLQKRGKEKLKKPNSLKSEVGAVTESVNQQVQITQGAKHLQTEDDISHT 248
M + +P +L+ VLQ+ GK+ ++ G + +N+ V+ ++G Q E D+ H
Sbjct: 887 MVQSNPQILQPVLQELGKQNPGLLRLIQEHHGEFLQLINEPVEGSEGDMFEQPEQDMPHA 1066
Query: 249 IMAPPSKKQ 257
I P++++
Sbjct: 1067INVTPAEQE 1093
>TC225670 UP|P93167 (P93167) Asparagine synthetase 2 , complete
Length = 2130
Score = 31.2 bits (69), Expect = 3.5
Identities = 16/63 (25%), Positives = 32/63 (50%), Gaps = 3/63 (4%)
Frame = +2
Query: 374 KSVDWEAVWTYALGPQPELALSLRM*LDDNHNSVVLACAKVV---QSALSCDVNENYFDI 430
K+V+W+A W+ L P AL + + +N N+ + K++ + L + Y D+
Sbjct: 1733 KAVEWDAAWSNNLDPSGRAALGVHISAYENQNNKGVEIEKIIPMDAAPLGVAIQG*YKDV 1912
Query: 431 SEN 433
++N
Sbjct: 1913 TKN 1921
>TC207076 similar to UP|Q8LAJ0 (Q8LAJ0) MutT domain protein-like, partial
(69%)
Length = 1502
Score = 30.0 bits (66), Expect = 7.9
Identities = 16/54 (29%), Positives = 27/54 (49%)
Frame = +2
Query: 610 LPLSIDDWLKLGKEKCKLKSALTIEQLRFWRVCIRYGYCVSHFSKIFPALCFWL 663
L LS+ W K+GK+ LK L +EQ + ++ G+ H + L +W+
Sbjct: 560 LRLSLSQWKKMGKKGIWLK--LPLEQSDLVPIAVKEGFQYHHAEPGYVMLTYWI 715
>TC218131 similar to UP|Q9LFZ9 (Q9LFZ9) F20N2.11, partial (8%)
Length = 1077
Score = 30.0 bits (66), Expect = 7.9
Identities = 25/82 (30%), Positives = 38/82 (45%), Gaps = 7/82 (8%)
Frame = -2
Query: 1256 KKHIEFLRFKSDIHESYSTFIEELVEQFSSISYGDLIFGRQVSVYLHCC-------VESS 1308
KK+ FL F + I S+ FI +Q S+G I +V L CC + S
Sbjct: 776 KKN*NFLSFSNAIQTSFFFFIFSPFKQTKGQSFGC*I*SSEVLTTLTCCRNLNLSTEQPS 597
Query: 1309 IRLATWNTLSNARVLELLPPLE 1330
+R A + TL ++L+ L L+
Sbjct: 596 LRDAIYLTLRCLQILQQLSNLK 531
>BG510825
Length = 440
Score = 30.0 bits (66), Expect = 7.9
Identities = 23/55 (41%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Frame = -2
Query: 904 EGIVSRCLVELRSM-LDVFTFSASSGWQRMQSIEIFGRGGPAPGMGVGWGAHGGG 957
EG+ S LVELR M L + GW R++ RGG G G G G GGG
Sbjct: 343 EGVCS-ALVELRKMRLLLLVGYGFHGWDRVRRF----RGGGGGGGGGGGGGGGGG 194
>TC219248 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich protein,
partial (11%)
Length = 450
Score = 30.0 bits (66), Expect = 7.9
Identities = 13/19 (68%), Positives = 13/19 (68%)
Frame = -2
Query: 939 GRGGPAPGMGVGWGAHGGG 957
G GGP PG G GWG GGG
Sbjct: 314 GPGGPGPG-GPGWGPGGGG 261
>TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferase like
protein, partial (27%)
Length = 820
Score = 30.0 bits (66), Expect = 7.9
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Frame = -2
Query: 1353 YAKSWVSDALDRAEIRGSVSYTMAVH-HLSSFIFNACPVDKL 1393
+A SW + +E SV+ ++A H HLS+F+ N+CP L
Sbjct: 603 FATSWRKVLILLSEEPPSVTASLAKHFHLSAFLLNSCPFPPL 478
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,818,801
Number of Sequences: 63676
Number of extensions: 931113
Number of successful extensions: 4998
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 4886
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4990
length of query: 1477
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1368
effective length of database: 5,698,948
effective search space: 7796160864
effective search space used: 7796160864
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC135101.10


