

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135100.7 + phase: 0 /pseudo
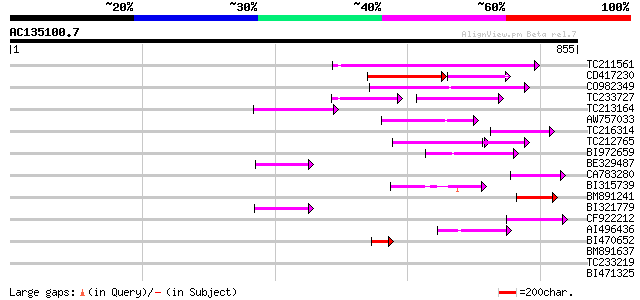
(855 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 162 4e-40
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 107 3e-34
CO982349 118 1e-26
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 64 1e-22
TC213164 78 2e-14
AW757033 74 3e-13
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 74 4e-13
TC212765 70 3e-12
BI972659 67 3e-11
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 66 7e-11
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 63 6e-10
BI315739 62 8e-10
BM891241 56 6e-08
BI321779 56 6e-08
CF922212 56 8e-08
AI496436 55 2e-07
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 43 5e-04
BM891637 42 0.002
TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance... 40 0.003
BI471325 36 0.063
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 162 bits (411), Expect = 4e-40
Identities = 102/313 (32%), Positives = 163/313 (51%)
Frame = +1
Query: 487 NAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLIMSC 546
+A+IA EV+ RS K L K+ EKAYD + W+FL + F I I C
Sbjct: 7 SALIANEVID--EAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 547 VKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHI 606
VK++ +++L NG+ T +F P +GLRQGDPL+P+LF + E L ++ VE+ +P +
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLV 360
Query: 607 SKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPE 666
+G +S L +ADD + F EA + V+ + L F S L++N KS V +
Sbjct: 361 GANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKS-CFGVFGVTD 537
Query: 667 RTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMHHRLASWKGKLLNKSGR 726
+ K++ L++ YLG P+ + II K +L+ WK + ++ GR
Sbjct: 538 QWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK*RHISFGGR 717
Query: 727 VCLEKSVTTSMPVYSMQINLLRKELCDGIDKITKSFIWGGNGVT*KWNMVKWTTVTSPRK 786
V L KSV TS+P+Y + + + D + KI ++F+WGG K + ++W + P++
Sbjct: 718 VTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRWEKLCLPKE 897
Query: 787 FGGLAIRDARNTN 799
GG+ R N N
Sbjct: 898 RGGI*KRLPNNHN 936
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 107 bits (267), Expect(2) = 3e-34
Identities = 52/119 (43%), Positives = 78/119 (64%)
Frame = -3
Query: 540 IGLIMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDR 599
+ L+ C+ + +++LWNG + F P G+RQ DP++PYLFVLC+E+L+ I + +
Sbjct: 686 VQLVWYCMSTTTMSLLWNGEVLDHFSPNIGVRQRDPIAPYLFVLCLERLSHLINLVMNKK 507
Query: 600 S*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKS 658
*R I +++ G +SHL F DD++LF EAN QV ++ TL F +SG KVNV K+KS
Sbjct: 506 L*RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLTLELFSDSSGAKVNVDKTKS 330
Score = 57.4 bits (137), Expect(2) = 3e-34
Identities = 34/95 (35%), Positives = 51/95 (52%)
Frame = -2
Query: 661 SKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMHHRLASWKGKL 720
SK V + DI ++ +LG YLG P+ T+ T+ II+K++ R + WK L
Sbjct: 321 SKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRFSRWKSNL 142
Query: 721 LNKSGRVCLEKSVTTSMPVYSMQINLLRKELCDGI 755
L+ GR+ L K V ++P + MQ + L LC GI
Sbjct: 141 LSMVGRLTLTK*VV*ALPSHIMQFDNL---LCMGI 46
>CO982349
Length = 795
Score = 118 bits (295), Expect = 1e-26
Identities = 72/242 (29%), Positives = 122/242 (49%)
Frame = +3
Query: 543 IMSCVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*R 602
I C+ ++ ++IL N + N+F P +GLRQGDPL+P LF + E L ++ V+ +
Sbjct: 72 IRGCLHSASVSILVNESPINEFSPQRGLRQGDPLAPLLFNIVAEGLTGLMREAVDRKRFN 251
Query: 603 PIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSK 662
+ K+ +S L +ADD + F EA + ++++ L F ASGLK+N +S+
Sbjct: 252 SFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKTILRCFELASGLKINFAESRFGAIW 431
Query: 663 RVPERTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMHHRLASWKGKLLN 722
+ P+ ++ + ++S YLG P+ E II K +LA WK + ++
Sbjct: 432 K-PDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRCREI*DPIIRKCETKLARWKQRHIS 608
Query: 723 KSGRVCLEKSVTTSMPVYSMQINLLRKELCDGIDKITKSFIWGGNGVT*KWNMVKWTTVT 782
GRV L ++ T++P+Y ++ + I + F+WGG + VKW TV
Sbjct: 609 LGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNIQRKFLWGGGSXQRRIAWVKWETVC 788
Query: 783 SP 784
P
Sbjct: 789 LP 794
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 63.5 bits (153), Expect(2) = 1e-22
Identities = 37/131 (28%), Positives = 66/131 (50%)
Frame = -2
Query: 614 SHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQ 673
SH+ ADD+++FC+ ++ V +I+ + + G ++ KSK +P + R I
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIGS-IPGQRLRVIS 275
Query: 674 GISSIRLVSNLGHYLGFPLVQGRVTNETYKHIIEKMHHRLASWKGKLLNKSGRVCLEKSV 733
+ +Y G P+ +G+ T + +K+ +LASWKG +L+ GR+ L SV
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 734 TTSMPVYSMQI 744
M +YS +
Sbjct: 94 IHGMLLYSFSV 62
Score = 62.4 bits (150), Expect(2) = 1e-22
Identities = 39/108 (36%), Positives = 59/108 (54%), Gaps = 1/108 (0%)
Frame = -3
Query: 486 DNAIIAQEVVHYMHTSRSQKGTLALKIYLEKAYDRLDWSFLELTLHDFGFPHHIIGLIMS 545
D + EV++ M + G LALKI + KA+D LD FL L+ FG+ H I
Sbjct: 828 DCTCVTSEVIN-MLDKKVFGGNLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRV 652
Query: 546 CVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPYLF-VLCMEKLAMHI 592
+ ++ L+I NG F +G+RQGDPLSP L+ C+E+ +++
Sbjct: 651 ILLSAKLSISVNGEPVGFFSCQRGVRQGDPLSPSLYRQRCLEQGTIYV 508
>TC213164
Length = 446
Score = 77.8 bits (190), Expect = 2e-14
Identities = 41/128 (32%), Positives = 67/128 (52%)
Frame = +3
Query: 368 KEEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVETLL 427
K+EV+ ++ + K+PGPDG F K FW+ L +D + G + L
Sbjct: 36 KKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKRSNAFFL 215
Query: 428 VLIPKENNPTHLKNFRPISLCNVIFKVITKVLVNRIRPFLDELVSPFQSNFIPRRGTTDN 487
LIPK ++P L ++PISL I+K++ K+L R + L ++ Q+ F+ R T N
Sbjct: 216 ALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEGRHTLHN 395
Query: 488 AIIAQEVV 495
+IA E++
Sbjct: 396 VVIANEIM 419
>AW757033
Length = 441
Score = 73.9 bits (180), Expect = 3e-13
Identities = 49/146 (33%), Positives = 75/146 (50%)
Frame = -2
Query: 561 TNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLTFAD 620
T +F P +GLRQGDPL+P LF + E L ++ V + + K +S L +AD
Sbjct: 440 TREFSPHRGLRQGDPLAPLLFNIAAEGLTGLMREAVARNHYKSFAVGKYKKPVSILQYAD 261
Query: 621 DVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQGISSIRL 680
D + F EA + V+++ L F ASGLK+N KS+ + VP+ +++ + L
Sbjct: 260 DTIFFGEATMENVRVIKTILRGFELASGLKINFAKSR-FGAISVPD*WRKEAAEFMNCSL 84
Query: 681 VSNLGHYLGFPLVQGRVTNETYKHII 706
+S YLG P+ ET+ II
Sbjct: 83 LSLPFSYLGIPIGANPRRRETWDPII 6
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 73.6 bits (179), Expect = 4e-13
Identities = 34/97 (35%), Positives = 55/97 (56%)
Frame = +2
Query: 725 GRVCLEKSVTTSMPVYSMQINLLRKELCDGIDKITKSFIWGGNGVT*KWNMVKWTTVTSP 784
GRV L KSV +++P+ + + + + D + + + F+WGG + VKW + +P
Sbjct: 8 GRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICNP 187
Query: 785 RKFGGLAIRDARNTNLALLGKLVWSLLHDKNKLWVRL 821
+ GGL I+D N N AL G+ +W L + N+LW RL
Sbjct: 188 KIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>TC212765
Length = 637
Score = 70.5 bits (171), Expect = 3e-12
Identities = 48/146 (32%), Positives = 69/146 (46%)
Frame = +3
Query: 578 PYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQMVI 637
PYLFVLCME+LA+ I RPI +S D + +L DD+ LF A +
Sbjct: 15 PYLFVLCMERLALCIHDLTSQ*IWRPILVSDDNRLVPYLMLVDDIFLFINAIGD*AYLAS 194
Query: 638 DTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQGRV 697
TL F GLK+N+ KS CS R+ + I +I +LG YL + GR
Sbjct: 195 LTL*NFFQTFGLKLNLDKSSMYCS*RMAPVLRDLISNTLNIVHAPSLGKYLCLKFIHGRS 374
Query: 698 TNETYKHIIEKMHHRLASWKGKLLNK 723
+ ++E++H+ +G L K
Sbjct: 375 KIVDFL-LVERIHNPWPRGRGSFLTK 449
Score = 57.4 bits (137), Expect = 3e-08
Identities = 27/72 (37%), Positives = 42/72 (57%)
Frame = +2
Query: 713 LASWKGKLLNKSGRVCLEKSVTTSMPVYSMQINLLRKELCDGIDKITKSFIWGGNGVT*K 772
LASWKGKLLN++ + CL V + P+Y M+ + L + + IDK ++ IW G +
Sbjct: 416 LASWKGKLLNQASKHCLT*LVLHAFPIYPMEASWLPQSIFQAIDKASRVTIWNKVGASLS 595
Query: 773 WNMVKWTTVTSP 784
W+ V +T + P
Sbjct: 596 WHSVS*STASKP 631
>BI972659
Length = 453
Score = 67.4 bits (163), Expect = 3e-11
Identities = 39/141 (27%), Positives = 69/141 (48%)
Frame = +1
Query: 627 EANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRDIQGISSIRLVSNLGH 686
EA+ + ++ L F SGL++N KS+ P D + + R + H
Sbjct: 7 EASWDNIIVLKSMLRGFEMVSGLRINYAKSQFGIVGFQPNWA-HDAAQLLNCRQLDIPFH 183
Query: 687 YLGFPLVQGRVTNETYKHIIEKMHHRLASWKGKLLNKSGRVCLEKSVTTSMPVYSMQINL 746
YLG P+ + ++ +I K +L+ W K L+ GRV L KSV ++P+Y +
Sbjct: 184 YLGMPIAVKASSRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFK 363
Query: 747 LRKELCDGIDKITKSFIWGGN 767
+ + + D + + ++F+WGGN
Sbjct: 364 IPQRIVDKLVSLQRTFMWGGN 426
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 65.9 bits (159), Expect = 7e-11
Identities = 30/88 (34%), Positives = 48/88 (54%)
Frame = -2
Query: 371 VKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVETLLVLI 430
+K A++ S K+PGPDG F KHFWE+ + ++ G + + + LI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 431 PKENNPTHLKNFRPISLCNVIFKVITKV 458
PK +P L +FRPISL ++K++ K+
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKI 112
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 62.8 bits (151), Expect = 6e-10
Identities = 31/84 (36%), Positives = 46/84 (53%)
Frame = +2
Query: 755 IDKITKSFIWGGNGVT*KWNMVKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDK 814
++ + + F+WGG + K V W TV P+ GGL I+D R N LLGK W L + +
Sbjct: 41 LESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTLLGKWRWDLFYIQ 220
Query: 815 NKLWVRLLSGKIY*EWLFMEQRSS 838
+ W ++L K Y W +E+ SS
Sbjct: 221 QEPWAKVLQSK-YGGWRALEEGSS 289
>BI315739
Length = 442
Score = 62.4 bits (150), Expect = 8e-10
Identities = 48/152 (31%), Positives = 76/152 (49%), Gaps = 7/152 (4%)
Frame = +3
Query: 575 PLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLTFADDVLLFCEANTQQVQ 634
PLSPYLF +CMEKLA+ IQ KV + + PI IS++GPG+S + + ++ F +
Sbjct: 48 PLSPYLF*ICMEKLAILIQSKVNEETWEPIKISRNGPGISPIYSSQIIVYFL------*K 209
Query: 635 MVIDTLN--EFCSASGLKVNVTKSKSMCSKRV-PERTKRDIQG----ISSIRLVSNLGHY 687
++ LN E C C+ V PE ++R + + L+ L
Sbjct: 210 PLVLKLNW*EMC---------------CNSFV*PEVSRRVFKNPGF*HKRMCLIEKLPGS 344
Query: 688 LGFPLVQGRVTNETYKHIIEKMHHRLASWKGK 719
L L+ GR E + H+++ +++RL WK K
Sbjct: 345 LL**LLAGRTKKEDFAHVVDMINNRLDGWKSK 440
>BM891241
Length = 407
Score = 56.2 bits (134), Expect = 6e-08
Identities = 26/61 (42%), Positives = 37/61 (60%)
Frame = -2
Query: 765 GGNGVT*KWNMVKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDKNKLWVRLLSG 824
GG+ K VKW V P+ GGL I+D N+ALLGK +W+L D+ +LW R+++
Sbjct: 406 GGDHDHKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
Query: 825 K 825
K
Sbjct: 226 K 224
>BI321779
Length = 421
Score = 56.2 bits (134), Expect = 6e-08
Identities = 28/89 (31%), Positives = 45/89 (50%)
Frame = +2
Query: 369 EEVKDALFSMQSDKAPGPDGFQPLFFKHFWEKTGESLWELVDISFRRGFIDEAIVETLLV 428
EE+K + +S K+PGPDG+ L K FWE E + + F+ + +
Sbjct: 104 EEIKKVI*DCESSKSPGPDGYSLLLIKIFWEVIKEEVINFFKEFHKFDFLPRGTNRSFIA 283
Query: 429 LIPKENNPTHLKNFRPISLCNVIFKVITK 457
LI K +NP + + PISL ++K++ K
Sbjct: 284 LIAKCDNPLDIGQYCPISLVGCLYKIV*K 370
>CF922212
Length = 445
Score = 55.8 bits (133), Expect = 8e-08
Identities = 31/91 (34%), Positives = 48/91 (52%)
Frame = -2
Query: 750 ELCDGIDKITKSFIWGGNGVT*KWNMVKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWS 809
++ D + ++ + F+WGG K V W +V P++ GL RD R N ALLGK W+
Sbjct: 432 KVLDKLVRLQQWFLWGGGLGQKKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKRRWN 253
Query: 810 LLHDKNKLWVRLLSGKIY*EWLFMEQRSSEK 840
L H + +L R+L K Y W +++ K
Sbjct: 252 LFHHQGELGARVLDSK-YKRWRNLDEERRVK 163
>AI496436
Length = 414
Score = 54.7 bits (130), Expect = 2e-07
Identities = 33/113 (29%), Positives = 59/113 (52%), Gaps = 2/113 (1%)
Frame = +1
Query: 646 ASGLKVNVTKSK--SMCSKRVPERTKRDIQGISSIRLVSNLGHYLGFPLVQGRVTNETYK 703
+SGLK+N KS+ ++C + + D S ++S YLG P+ ++ ++
Sbjct: 25 SSGLKINFAKSQFGTICKSDIWRKEAADYLNSS---VLSMPFVYLGIPIGANSRHSDVWE 195
Query: 704 HIIEKMHHRLASWKGKLLNKSGRVCLEKSVTTSMPVYSMQINLLRKELCDGID 756
++ K +LASWK K ++ GRV L SV T++P+Y + + ++ D D
Sbjct: 196 PVVRKCERKLASWKQKYVSFGGRVTLINSVLTALPIYLLSFFRIPDKVVDKAD 354
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 43.1 bits (100), Expect = 5e-04
Identities = 16/34 (47%), Positives = 25/34 (73%)
Frame = +1
Query: 546 CVKASDLAILWNGAKTNKFKPTKGLRQGDPLSPY 579
C+ ++ ++IL NG+ T +FKP +GLRQG P P+
Sbjct: 7 CLSSASISILVNGSPTEEFKPXRGLRQGGPFGPF 108
>BM891637
Length = 427
Score = 41.6 bits (96), Expect = 0.002
Identities = 23/68 (33%), Positives = 34/68 (49%)
Frame = -1
Query: 776 VKWTTVTSPRKFGGLAIRDARNTNLALLGKLVWSLLHDKNKLWVRLLSGKIY*EWLFMEQ 835
+ W + GGL I+D + N ALL K W + H ++LW R+L K Y W ++Q
Sbjct: 403 ISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRILISK-YKGWRGLDQ 227
Query: 836 RSSEKPFS 843
+ FS
Sbjct: 226 GPQKYYFS 203
>TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance
protein-like protein (Fragment), partial (74%)
Length = 781
Score = 40.4 bits (93), Expect = 0.003
Identities = 29/102 (28%), Positives = 48/102 (46%)
Frame = +3
Query: 558 GAKTNKFKPTKGLRQGDPLSPYLFVLCMEKLAMHIQGKVEDRS*RPIHISKDGPGLSHLT 617
G +++ F T GL QG LSPYLF L ++ L IQ V +
Sbjct: 99 GGESDDFPITIGLHQGSTLSPYLFTLILDVLTEQIQEIVS----------------RCML 230
Query: 618 FADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSM 659
FADD++L E+ +++ ++T G +++ +KS+ M
Sbjct: 231 FADDIVLLGESR-EKLNERLETWRRALETHGFRLSRSKSEYM 353
>BI471325
Length = 421
Score = 36.2 bits (82), Expect = 0.063
Identities = 21/59 (35%), Positives = 32/59 (53%)
Frame = +2
Query: 613 LSHLTFADDVLLFCEANTQQVQMVIDTLNEFCSASGLKVNVTKSKSMCSKRVPERTKRD 671
LSHL D LFC ++ ++ + L F ++SGL VN+ KS+ +C R + RD
Sbjct: 44 LSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSE-ICFSRNTSQIARD 217
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.345 0.152 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,683,510
Number of Sequences: 63676
Number of extensions: 570372
Number of successful extensions: 5009
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 3680
Number of HSP's successfully gapped in prelim test: 141
Number of HSP's that attempted gapping in prelim test: 1269
Number of HSP's gapped (non-prelim): 3913
length of query: 855
length of database: 12,639,632
effective HSP length: 105
effective length of query: 750
effective length of database: 5,953,652
effective search space: 4465239000
effective search space used: 4465239000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135100.7


