

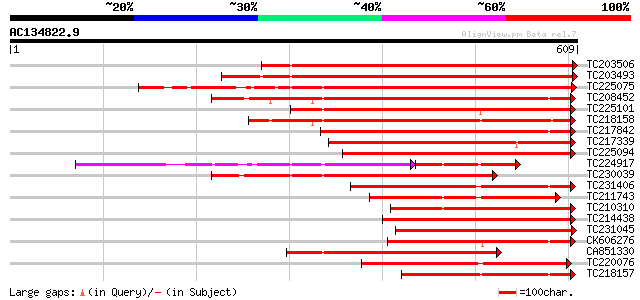
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.9 + phase: 0
(609 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 625 e-179
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 613 e-176
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 423 e-118
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 361 e-100
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 360 1e-99
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 356 1e-98
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 345 3e-95
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 335 3e-92
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 318 3e-87
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 196 1e-85
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 311 7e-85
TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (... 275 3e-74
TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 273 2e-73
TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 273 2e-73
TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (F... 254 6e-68
TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase iso... 254 6e-68
CK606276 245 5e-65
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 242 4e-64
TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 pr... 240 2e-63
TC218157 similar to UP|Q8GUF8 (Q8GUF8) At2g47550/T30B22.15, part... 240 2e-63
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 625 bits (1613), Expect = e-179
Identities = 297/339 (87%), Positives = 320/339 (93%)
Frame = +2
Query: 271 EFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRA 330
+FP WL KRDRRLLSLP+S IQADI VSK+G NGTVKTI EAIKKAPEHS RRFIIYVRA
Sbjct: 8 KFPGWLNKRDRRLLSLPLSAIQADITVSKTG-NGTVKTIAEAIKKAPEHSTRRFIIYVRA 184
Query: 331 GRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDIT 390
GRYEENNLKVG+KKTN+MFIGDG+GKTVITGK++V DGMTTFH+ASFAASG GF+ARDIT
Sbjct: 185 GRYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNVIDGMTTFHSASFAASGAGFIARDIT 364
Query: 391 FENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGN 450
FENYAGP KHQAVALRVG+DHAVVYRCNIVGYQD+CYVHSNRQFFRECNIYGTVDFIFGN
Sbjct: 365 FENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIFGN 544
Query: 451 AAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETY 510
AAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGIS+H+CRILPAPDLA KGS TY
Sbjct: 545 AAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISLHNCRILPAPDLAPVKGSFPTY 724
Query: 511 LGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWP 570
LGRPWK YSRTVY++SYMGDH+HP GWLEWNGDFAL TLYYGEYMN+GPGAA+GQRV+WP
Sbjct: 725 LGRPWKQYSRTVYLVSYMGDHIHPRGWLEWNGDFALNTLYYGEYMNYGPGAAVGQRVQWP 904
Query: 571 GYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGLST 609
GYRVI ST+EANR+TVAQFISGS+WLPSTGVAF AGLST
Sbjct: 905 GYRVIKSTMEANRFTVAQFISGSAWLPSTGVAFAAGLST 1021
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 613 bits (1582), Expect = e-176
Identities = 297/384 (77%), Positives = 338/384 (87%), Gaps = 2/384 (0%)
Frame = +1
Query: 228 NLKDLSELVSNSLAIFSASG-DNDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRRLLSL 286
NLKDLSELVSN LAIFS +G +DF GVPIQN+RRLM M + + FP WL RDRRLLSL
Sbjct: 10 NLKDLSELVSNCLAIFSGAGAGDDFAGVPIQNRRRLMAMREDN--FPTWLNGRDRRLLSL 183
Query: 287 PVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTN 346
P+S+IQADI+VSK G NGTVKTI EAIKK PE+S RR IIY+RAGRYEE+NLK+G+KKTN
Sbjct: 184 PLSQIQADIVVSKDG-NGTVKTIAEAIKKVPEYSSRRIIIYIRAGRYEEDNLKLGRKKTN 360
Query: 347 IMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALR 406
+MFIGDG+GKTVITG R+ +TTFHTASFAASG GF+A+D+TFENYAGP +HQAVALR
Sbjct: 361 VMFIGDGKGKTVITGGRNYYQNLTTFHTASFAASGSGFIAKDMTFENYAGPGRHQAVALR 540
Query: 407 VGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPM 466
VG+DHAVVYRCNI+GYQD YVHSNRQF+REC+IYGTVDFIFGNAAVVFQ C ++ARKPM
Sbjct: 541 VGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFIFGNAAVVFQNCTLWARKPM 720
Query: 467 AQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLS 526
AQQKNTITAQNRKDPNQNTGISIH+CRI+ PDL +SKGS TYLGRPWK+Y+RTV+MLS
Sbjct: 721 AQQKNTITAQNRKDPNQNTGISIHNCRIMATPDLEASKGSYPTYLGRPWKLYARTVFMLS 900
Query: 527 YMGDHVHPHGWLEWN-GDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYT 585
Y+GDHVHP GWLEWN FAL T YYGEYMN+GPG+A+GQRV W GYR I ST+EA+R+T
Sbjct: 901 YIGDHVHPRGWLEWNTSSFALDTCYYGEYMNYGPGSALGQRVNWAGYRAINSTVEASRFT 1080
Query: 586 VAQFISGSSWLPSTGVAFLAGLST 609
V QFISGSSWLPSTGVAF+AGLST
Sbjct: 1081VGQFISGSSWLPSTGVAFIAGLST 1152
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 423 bits (1088), Expect = e-118
Identities = 222/470 (47%), Positives = 297/470 (62%)
Frame = +1
Query: 139 VANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMT 198
+AN R+ A DCL+L+D S D + ++ S N G+ N++ D+ T
Sbjct: 226 LANFRLSTAIADCLDLLDLSSDVLSWAL-------SASQNPKGKHNSTGNLSS---DLRT 375
Query: 199 WLSAALTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQN 258
WLSAAL + +TC+EGFE T+ VK + + + LV LA +P Q+
Sbjct: 376 WLSAALAHPETCMEGFEGTNSIVKGLVSAGIGQVVSLVEQLLAQV----------LPAQD 525
Query: 259 KRRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPE 318
+ + +FP W++ ++R+LL + D+ V+ G +G I +A+ AP+
Sbjct: 526 Q---FDAASSKGQFPSWIKPKERKLLQ--AIAVTPDVTVALDG-SGNYAKIMDAVLAAPD 687
Query: 319 HSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFA 378
+S +RF+I V+ G Y EN +++ KKK NIM +G G TVI+G RSV DG TTF +A+FA
Sbjct: 688 YSMKRFVILVKKGVYVEN-VEIKKKKWNIMILGQGMDATVISGNRSVVDGWTTFRSATFA 864
Query: 379 ASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFREC 438
SG GF+ARDI+F+N AGPEKHQAVALR SD +V +RC I GYQD+ Y H+ RQFFR+C
Sbjct: 865 VSGRGFIARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDC 1044
Query: 439 NIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAP 498
I GTVD+IFG+A VFQ C + +K + QKNTITA RKDPN+ TG S C I
Sbjct: 1045TISGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKDPNEPTGFSFQFCNITADS 1224
Query: 499 DLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFG 558
DL S G+ +TYLGRPWK YSRTV+M SYM + + GWLEWNG+FAL TLYY EYMN G
Sbjct: 1225DLIPSVGTAQTYLGRPWKSYSRTVFMQSYMSEVIGAEGWLEWNGNFALDTLYYAEYMNTG 1404
Query: 559 PGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGLS 608
GA + RVKWPGY + + +A+ +TV+QFI G+ WLPSTGV F AGL+
Sbjct: 1405AGAGVANRVKWPGYHALNDSSQASNFTVSQFIEGNLWLPSTGVTFTAGLT 1554
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 361 bits (926), Expect = e-100
Identities = 192/415 (46%), Positives = 260/415 (62%), Gaps = 24/415 (5%)
Frame = +1
Query: 217 TSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQNKRRLMGMSDISREFPKWL 276
T + + + LKD+++L S SL + + + D + +NK R G+ + + + L
Sbjct: 4 TKSNIANALAVPLKDVTQLYSVSLGLVTEALDKNLR----RNKTRKHGLPTKTFKVRQPL 171
Query: 277 EK-------------------RDRRLLSLPVSE--IQADIIVSKSGGNGTVKTITEAIKK 315
EK R R+L S+ + D ++ G +I +AI
Sbjct: 172 EKLIKLLRTKYSCAKLSNCTSRTERILKESGSQGILLYDFVIVSHYGIDNYTSIGDAIAA 351
Query: 316 APEHSRRR---FIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTF 372
AP +++ F++YVR G YEE + + K+K NI+ +GDG KT+ITG SV DG TTF
Sbjct: 352 APNNTKPEDGYFLVYVREGLYEEY-VVIPKEKKNILLVGDGINKTIITGNHSVIDGWTTF 528
Query: 373 HTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNR 432
++++FA SG F+A D+TF N AGPEKHQAVA+R +D + YRC+ GYQD YVHS R
Sbjct: 529 NSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAVRNNADLSTFYRCSFEGYQDTLYVHSLR 708
Query: 433 QFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDC 492
QF+REC IYGTVDFIFGNAAVVFQ C IYARKP+ QKN +TAQ R DPNQNTGISI +C
Sbjct: 709 QFYRECEIYGTVDFIFGNAAVVFQGCKIYARKPLPNQKNAVTAQGRTDPNQNTGISIQNC 888
Query: 493 RILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYG 552
I APDL + S ++LGRPWK+YSRTVY+ SY+G+ + P GWLEWNG L TL+YG
Sbjct: 889 SIDAAPDLVADLNSTMSFLGRPWKVYSRTVYLQSYIGNVIQPAGWLEWNGTVGLDTLFYG 1068
Query: 553 EYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
E+ N+GPG+ RV WPGY ++ +T +A +TV F G++WLP T + + GL
Sbjct: 1069EFNNYGPGSNTSNRVTWPGYSLLNAT-QAWNFTVLNFTLGNTWLPDTDIPYTEGL 1230
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 360 bits (923), Expect = 1e-99
Identities = 173/309 (55%), Positives = 221/309 (70%), Gaps = 3/309 (0%)
Frame = +3
Query: 302 GNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITG 361
G + +A+ AP +S +R++I+++ G Y EN +++ KKK N+M +GDG T+I+G
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYEN-VEIKKKKWNLMMVGDGMDATIISG 383
Query: 362 KRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVG 421
RS DG TTF +A+FA SG GF+ARDITF+N AGPEKHQAVALR SD +V +RC I G
Sbjct: 384 NRSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFG 563
Query: 422 YQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDP 481
YQD+ Y H+ RQF+REC I GTVDFIFG+A +FQ C+I A+K + QKNTITA RK+P
Sbjct: 564 YQDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAHGRKNP 743
Query: 482 NQNTGISIHDCRILPAPDLASSK---GSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWL 538
++ TG SI C I DL +S S TYLGRPWK YSRT++M SY+ D + P GWL
Sbjct: 744 DEPTGFSIQFCNISADYDLVNSVNSCNSTHTYLGRPWKPYSRTIFMQSYISDVLRPEGWL 923
Query: 539 EWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPS 598
EWNGDFAL TLYY EYMN+GPGA + RVKW GY V+ + +A+ +TV+QFI G+ LPS
Sbjct: 924 EWNGDFALDTLYYAEYMNYGPGAGVANRVKWQGYHVMNDSSQASNFTVSQFIEGNLCLPS 1103
Query: 599 TGVAFLAGL 607
TGV F AGL
Sbjct: 1104TGVTFTAGL 1130
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 356 bits (914), Expect = 1e-98
Identities = 189/356 (53%), Positives = 233/356 (65%), Gaps = 5/356 (1%)
Frame = +3
Query: 257 QNKRRLMGMSDISREFPKWLEKRDRRLLSLPVSE--IQADIIVSKSGGNGTVKTITEAIK 314
+N R + MS +R + + +R +LL V + + DI+ G+G TI +AI
Sbjct: 36 KNGRLPLKMSSRTRAIYESVSRR--KLLQAKVGDEVVVRDIVTVSQDGSGNFTTINDAIA 209
Query: 315 KAPEHSRRR---FIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTT 371
AP S F+IYV AG YEEN + + KKKT +M +GDG KT+ITG RSV DG TT
Sbjct: 210 AAPNKSVSTDGYFLIYVTAGVYEEN-VSIDKKKTYLMMVGDGINKTIITGNRSVVDGWTT 386
Query: 372 FHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSN 431
F +A+ A G GF+ ++T N AG KHQAVALR G+D + Y C+ GYQD YVHS
Sbjct: 387 FSSATLAVVGQGFVGVNMTIRNTAGAVKHQAVALRSGADLSTFYSCSFEGYQDTLYVHSL 566
Query: 432 RQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHD 491
RQF+ EC+IYGTVDFIFGNA VVFQ C +Y R PM+ Q N ITAQ R DPNQ+TGISIH+
Sbjct: 567 RQFYSECDIYGTVDFIFGNAKVVFQNCKMYPRLPMSGQFNAITAQGRTDPNQDTGISIHN 746
Query: 492 CRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYY 551
C I A DLA+S G + TYLGRPWK YSRTVYM + M +H GW EW+GDFAL TLYY
Sbjct: 747 CTIRAADDLAASNG-VATYLGRPWKEYSRTVYMQTVMDSVIHAKGWREWDGDFALSTLYY 923
Query: 552 GEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
EY N GPG+ RV WPGY VI +T AN +TV+ F+ G WLP TGV++ L
Sbjct: 924 AEYSNSGPGSGTDNRVTWPGYHVINATDAAN-FTVSNFLLGDDWLPQTGVSYTNNL 1088
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 345 bits (886), Expect = 3e-95
Identities = 159/274 (58%), Positives = 204/274 (74%)
Frame = +3
Query: 334 EENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFEN 393
E+ N+ + + N+M +GDG G T++TG + DG TTF +A+FA G GF+ARDITFEN
Sbjct: 3 EKENIDIKRTVKNLMIVGDGMGATIVTGNHNAIDGSTTFRSATFAVDGDGFIARDITFEN 182
Query: 394 YAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAV 453
AGP+KHQAVALR G+DH+V YRC+ GYQD YV++NRQF+R+C+IYGTVDFIFG+A
Sbjct: 183 TAGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDAVA 362
Query: 454 VFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGR 513
V Q CNIY RKPM+ Q+NT+TAQ R DPN+NTGI IH+CRI A DL + +GS T+LGR
Sbjct: 363 VLQNCNIYVRKPMSNQQNTVTAQGRTDPNENTGIIIHNCRITAAGDLKAVQGSFRTFLGR 542
Query: 514 PWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYR 573
PW+ YSRTV M S + + P GW W+G+FAL TLYY E+ N G GA+ G RV W G+R
Sbjct: 543 PWQKYSRTVVMKSALDGLISPAGWFPWSGNFALSTLYYAEHANTGAGASTGGRVDWAGFR 722
Query: 574 VITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
VI+ST EA ++TV F++G SW+P +GV F GL
Sbjct: 723 VISST-EAVKFTVGNFLAGGSWIPGSGVPFDEGL 821
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 335 bits (859), Expect = 3e-92
Identities = 161/268 (60%), Positives = 199/268 (74%), Gaps = 3/268 (1%)
Frame = +1
Query: 343 KKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQA 402
+KTN+M +GDG+ TVITG + DG TTF TA+ AA G GF+A+DI F+N AGP+KHQA
Sbjct: 1 EKTNVMLVGDGKDATVITGNLNFIDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQA 180
Query: 403 VALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYA 462
VALRVG+D +V+ RC I +QD Y HSNRQF+R+ I GTVDFIFGNAAVVFQKC++ A
Sbjct: 181 VALRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVA 360
Query: 463 RKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTV 522
RKPM +Q N +TAQ R+DPNQNTG SI C + P+ DL GSI+T+LGRPWK YSRTV
Sbjct: 361 RKPMDKQNNMVTAQGREDPNQNTGTSIQQCNLTPSSDLKPVVGSIKTFLGRPWKKYSRTV 540
Query: 523 YMLSYMGDHVHPHGWLEWNG---DFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTL 579
M S + H+ P GW EW+ DF L+TLYYGEYMN GPGA +RV WPGY +I +
Sbjct: 541 VMQSTLDSHIDPTGWAEWDAQSKDF-LQTLYYGEYMNNGPGAGTSKRVNWPGYHIIKTAA 717
Query: 580 EANRYTVAQFISGSSWLPSTGVAFLAGL 607
EA+++TVAQ I G+ WL +TGV F+ GL
Sbjct: 718 EASKFTVAQLIQGNVWLKNTGVNFIEGL 801
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 318 bits (816), Expect = 3e-87
Identities = 150/250 (60%), Positives = 182/250 (72%)
Frame = +3
Query: 358 VITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRC 417
+I+G RSV DG TTF +A+FA SG GF+ARDI+F+N AGPEKHQAVALR +D +V +RC
Sbjct: 3 IISGNRSVVDGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRC 182
Query: 418 NIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQN 477
I GYQD+ Y H+ RQFFREC I GTVD+IFG+A VFQ C + +K + QKNTITA
Sbjct: 183 GIFGYQDSLYTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHG 362
Query: 478 RKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGW 537
RKDPN+ TG S C I DL S ++YLGRPWK YSRTV+M SYM + + GW
Sbjct: 363 RKDPNEPTGFSFQFCNITADSDLVPWVSSTQSYLGRPWKSYSRTVFMQSYMSEVIRGEGW 542
Query: 538 LEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLP 597
LEWNG+FAL+TLYYGEYMN G GA + RVKWPGY + +A+ +TVAQFI G+ WLP
Sbjct: 543 LEWNGNFALETLYYGEYMNTGAGAGLANRVKWPGYHPFNDSNQASNFTVAQFIEGNLWLP 722
Query: 598 STGVAFLAGL 607
STGV + AGL
Sbjct: 723 STGVTYTAGL 752
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 196 bits (498), Expect(2) = 1e-85
Identities = 125/368 (33%), Positives = 190/368 (50%), Gaps = 2/368 (0%)
Frame = +2
Query: 71 SNLLHSKPTQAISRTCSKTRYPSLCINSLLDFP-GSTSASEQELVHISFNMTHRHISKAL 129
S++ S + I C++T YP C L + S+ + + +S + ++
Sbjct: 167 SSIASSYSFKDIQSWCNQTPYPQPCEYYLTNHAFNKPIKSKSDFLKVSLQLALERAQRSE 346
Query: 130 FASSGLSYTVANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNV 189
+ L N +AA+ DCL+L + ++ + +++ N
Sbjct: 347 LNTHALGPKCRNVHEKAAWADCLQLYEYTIQRLNKTINP-------------------NT 469
Query: 190 AGSTEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQMVGNLKD-LSELVSNSLAIFSASGD 248
+ D TWLS ALTN +TC GF + V D ++ + + +++L+SN+L++
Sbjct: 470 KCNQTDTQTWLSTALTNLETCKNGFYELG--VPDYVLPLMSNNVTKLLSNTLSLNKG--- 634
Query: 249 NDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKT 308
P Q K FP W++ DR+LL A+++V+K G +G T
Sbjct: 635 ------PYQYKP-----PSYKEGFPTWVKPGDRKLLQSSSVASNANVVVAKDG-SGKYTT 778
Query: 309 ITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDG 368
+ A+ AP+ S R++IYV++G Y E +V K NIM +GDG GKT+ITG +SVG G
Sbjct: 779 VKAAVDAAPKSSSGRYVIYVKSGVYNE---QVEVKGNNIMLVGDGIGKTIITGSKSVGGG 949
Query: 369 MTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYV 428
TTF +A+ AA G GF+A+DITF N AG HQAVA R GSD +V YRC+ G+QD YV
Sbjct: 950 TTTFRSATVAAVGDGFIAQDITFRNTAGAANHQAVAFRSGSDLSVFYRCSFEGFQDTLYV 1129
Query: 429 HSNRQFFR 436
HS RQF+R
Sbjct: 1130HSERQFYR 1153
Score = 139 bits (351), Expect(2) = 1e-85
Identities = 66/112 (58%), Positives = 84/112 (74%)
Frame = +3
Query: 437 ECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILP 496
EC+IYGTVDFIFGNAA V Q CNIYAR P Q+ T+TAQ R DPNQNTGI IH+ ++
Sbjct: 1155 ECDIYGTVDFIFGNAAAVLQNCNIYARTP-PQRTITVTAQGRTDPNQNTGIIIHNSKVTG 1331
Query: 497 APDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKT 548
A S S+++YLGRPW+ YSRTV+M +Y+ ++P GW+EW+G+FAL T
Sbjct: 1332 ASGFNPS--SVKSYLGRPWQKYSRTVFMKTYLDSLINPAGWMEWDGNFALDT 1481
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 311 bits (796), Expect = 7e-85
Identities = 161/310 (51%), Positives = 208/310 (66%), Gaps = 2/310 (0%)
Frame = +1
Query: 217 TSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVP-IQNKRRLM-GMSDISREFPK 274
+ G V+D+ L ++S VSNSLA+ GV + +K + G I FP
Sbjct: 1 SKGHVRDRFEEGLLEISHHVSNSLAMLKKLP----AGVKKLASKNEVFPGYGKIKDGFPT 168
Query: 275 WLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYE 334
WL +DR+LL V+E +++V+K G G TI EA+ AP S RF+I+++AG Y
Sbjct: 169 WLSTKDRKLLQAAVNETNFNLLVAKDG-TGNFTTIAEAVAVAPNSSATRFVIHIKAGAYF 345
Query: 335 ENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENY 394
EN ++V +KKTN+MF+GDG GKTV+ R+V DG TTF +A+ A G GF+A+ ITFEN
Sbjct: 346 EN-VEVIRKKTNLMFVGDGIGKTVVKASRNVVDGWTTFQSATVAVVGDGFIAKGITFENS 522
Query: 395 AGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVV 454
AGP KHQAVALR GSD + Y+C+ V YQD YVHS RQF+R+C++YGTVDFIFGNAA V
Sbjct: 523 AGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYVHSLRQFYRDCDVYGTVDFIFGNAATV 702
Query: 455 FQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRP 514
Q CN+YARKP Q+N TAQ R+DPNQNTGISI +C++ A DL K + YLGRP
Sbjct: 703 LQNCNLYARKPNENQRNLFTAQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGRP 882
Query: 515 WKMYSRTVYM 524
WK YSRTVY+
Sbjct: 883 WKKYSRTVYL 912
>TC231406 similar to UP|Q9FF78 (Q9FF78) Pectinesterase, partial (35%)
Length = 851
Score = 275 bits (704), Expect = 3e-74
Identities = 134/241 (55%), Positives = 166/241 (68%)
Frame = +2
Query: 367 DGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDAC 426
DG TF TA+FA G F+ARD+ F N AGP+K QAVAL +D AV YRC I +QD+
Sbjct: 8 DGTPTFSTATFAVFGRNFIARDMGFRNTAGPQKQQAVALMTSADQAVYYRCQIDAFQDSL 187
Query: 427 YVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTG 486
Y HSNRQF+RECNIYGTVDFIFGN+AVV Q CNI R PM Q+N ITAQ + DPN NTG
Sbjct: 188 YAHSNRQFYRECNIYGTVDFIFGNSAVVLQNCNIMPRVPMQGQQNPITAQGKTDPNMNTG 367
Query: 487 ISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFAL 546
ISI +C I P DL+ S++TYLGRPWK YS V+M S MG +HP+GWL W G+ A
Sbjct: 368 ISIQNCNITPFGDLS----SVKTYLGRPWKNYSTPVFM*SPMGIFIHPNGWLPWVGNSAP 535
Query: 547 KTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAG 606
T++Y E+ N GPGA+ RV W G RVIT +A+ +TV F+SG W+ ++G F +
Sbjct: 536 DTIFYAEFQNVGPGASTKNRVNWKGLRVITRK-QASMFTVKAFLSGERWITASGAPFKSS 712
Query: 607 L 607
+
Sbjct: 713 I 715
>TC211743 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (39%)
Length = 601
Score = 273 bits (698), Expect = 2e-73
Identities = 130/205 (63%), Positives = 156/205 (75%)
Frame = +2
Query: 387 RDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDF 446
+DITF N AG HQAVALR GSD +V YRC+ GYQD YV+S+RQF+REC+IYGTVDF
Sbjct: 2 QDITFRNTAGATNHQAVALRSGSDLSVFYRCSFEGYQDTLYVYSDRQFYRECDIYGTVDF 181
Query: 447 IFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGS 506
IFGNAAVVFQ CNIYAR P + NTITAQ R DPNQNTGISIH+ ++ A DL
Sbjct: 182 IFGNAAVVFQNCNIYARNP-PNKVNTITAQGRTDPNQNTGISIHNSKVTAASDLM----G 346
Query: 507 IETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQR 566
+ TYLGRPW+ YSRTV+M +Y+ ++P GWLEW+G+FAL TLYYGEYMN GPG++ R
Sbjct: 347 VRTYLGRPWQQYSRTVFMKTYLDSLINPEGWLEWSGNFALSTLYYGEYMNTGPGSSTANR 526
Query: 567 VKWPGYRVITSTLEANRYTVAQFIS 591
V W GY VITS EA+++TV FI+
Sbjct: 527 VNWLGYHVITSASEASKFTVGNFIA 601
>TC210310 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (38%)
Length = 678
Score = 273 bits (698), Expect = 2e-73
Identities = 126/198 (63%), Positives = 154/198 (77%)
Frame = +1
Query: 410 DHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQ 469
D +V Y+C+ GYQD YVHS RQF+RECNIYGTVDFIFGNAAVV Q CNI+AR P +
Sbjct: 1 DLSVFYKCSFEGYQDTLYVHSERQFYRECNIYGTVDFIFGNAAVVLQNCNIFARNP-PNK 177
Query: 470 KNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMG 529
NTITAQ R DPNQNTGISIH+ R+ A DL + S+ TYLGRPWK YSRTV+M +Y+
Sbjct: 178 VNTITAQGRTDPNQNTGISIHNSRVTAASDLRPVQNSVRTYLGRPWKQYSRTVFMKTYLD 357
Query: 530 DHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQF 589
++P GW+EW+G+FAL TLYYGEYMN GPG++ +RVKW GYRVITS EA++++VA F
Sbjct: 358 GLINPAGWMEWSGNFALDTLYYGEYMNTGPGSSTARRVKWSGYRVITSASEASKFSVANF 537
Query: 590 ISGSSWLPSTGVAFLAGL 607
I+G++WLPST V F L
Sbjct: 538 IAGNAWLPSTKVPFTPSL 591
>TC214438 similar to UP|Q708X4 (Q708X4) Pectin methylesterase (Fragment) ,
partial (49%)
Length = 1026
Score = 254 bits (650), Expect = 6e-68
Identities = 125/207 (60%), Positives = 144/207 (69%)
Frame = +3
Query: 401 QAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNI 460
QAVALRVG D + + C+I+ +QD YVH+NRQFF +C I GTVDFIFGN+AVVFQ C+I
Sbjct: 6 QAVALRVGGDLSAFFNCDILAFQDTLYVHNNRQFFEKCLIAGTVDFIFGNSAVVFQDCDI 185
Query: 461 YARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSR 520
+AR P + QKN +TAQ R DPNQNTGI I CRI DL S K + +TYLGRPWK YSR
Sbjct: 186 HARLPSSGQKNMVTAQGRVDPNQNTGIVIQKCRIGATNDLESVKKNFKTYLGRPWKEYSR 365
Query: 521 TVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLE 580
TV M S + D + P GW EW+G+FAL TL Y EY N GPGA RV W GY+VIT E
Sbjct: 366 TVIMQSSISDVIDPIGWHEWSGNFALSTLVYREYQNTGPGAGTSNRVTWKGYKVITDAAE 545
Query: 581 ANRYTVAQFISGSSWLPSTGVAFLAGL 607
A YT FI GSSWL STG F GL
Sbjct: 546 ARDYTPGSFIGGSSWLGSTGFPFSLGL 626
>TC231045 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (69%)
Length = 747
Score = 254 bits (650), Expect = 6e-68
Identities = 116/194 (59%), Positives = 137/194 (69%)
Frame = +2
Query: 415 YRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTIT 474
Y+C V YQD VHS RQF+R+C++YGTVDF GNAA V Q CN+YARKP Q+N T
Sbjct: 2 YKCXFVXYQDXXXVHSXRQFYRDCDVYGTVDFXXGNAATVLQNCNLYARKPNENQRNLFT 181
Query: 475 AQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHP 534
AQ R+DPNQNTGISI +C++ A DL K + YLGRPWK YSRTVY+ SYM D + P
Sbjct: 182 AQGREDPNQNTGISILNCKVAAAADLIPVKSQFKNYLGRPWKKYSRTVYLNSYMEDLIDP 361
Query: 535 HGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSS 594
GWLEWNG FAL TLYYGEY N GPG+ RV WPGYRVI + EAN++TV FI G+
Sbjct: 362 KGWLEWNGTFALDTLYYGEYNNRGPGSNTSARVTWPGYRVIKNATEANQFTVRNFIQGNE 541
Query: 595 WLPSTGVAFLAGLS 608
WL ST + F + S
Sbjct: 542 WLSSTDIPFFSDFS 583
>CK606276
Length = 751
Score = 245 bits (625), Expect = 5e-65
Identities = 117/205 (57%), Positives = 142/205 (69%), Gaps = 3/205 (1%)
Frame = +1
Query: 406 RVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKP 465
R G+D + Y C+ GYQD YVHS RQF++ C+IYGTVDFIFGNAA + Q CN+Y R P
Sbjct: 1 RNGADMSTFYNCSFEGYQDTLYVHSLRQFYKSCDIYGTVDFIFGNAAALLQDCNMYPRLP 180
Query: 466 MAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGS---IETYLGRPWKMYSRTV 522
M Q N ITAQ R DPNQNTGISI +C I+ A DL + + I+TYLGRPWK YSRTV
Sbjct: 181 MQNQFNAITAQGRTDPNQNTGISIQNCCIIAASDLGDATNNYNGIKTYLGRPWKEYSRTV 360
Query: 523 YMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEAN 582
YM S++ + P GW EW+GDFAL TLYY E+ N+GPG+ RV W GY +I +A+
Sbjct: 361 YMQSFIDGLIDPKGWNEWSGDFALSTLYYAEFANWGPGSNTSNRVAWEGYHLIDEK-DAD 537
Query: 583 RYTVAQFISGSSWLPSTGVAFLAGL 607
+TV +FI G WLP TGV F AGL
Sbjct: 538 DFTVHKFIQGEKWLPQTGVPFXAGL 612
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 242 bits (617), Expect = 4e-64
Identities = 123/232 (53%), Positives = 157/232 (67%), Gaps = 1/232 (0%)
Frame = +2
Query: 298 SKSGGNGTVKTITEAIKKAPEHSRR-RFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGK 356
++ G+G KT+ +A+ A + + RF+I+V+ G Y EN ++V NIM +GDG
Sbjct: 2 ARKXGSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYREN-IEVALHNDNIMLVGDGLRN 178
Query: 357 TVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYR 416
T+IT RSV DG TT+ +A+ G F+ARDITF+N AG K QAVALR SD +V YR
Sbjct: 179 TIITSARSVQDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYR 358
Query: 417 CNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQ 476
C I+GYQD H+ RQF+R+C IYGTVDFIFGNAAVVFQ C I+AR+P+ Q N ITAQ
Sbjct: 359 CGIMGYQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQ 538
Query: 477 NRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYM 528
R DP QNTGISIH+ +I APDL T+LGRPW+ YSR V M ++M
Sbjct: 539 GRGDPFQNTGISIHNSQIRAAPDLKPVVDKYNTFLGRPWQQYSRVVVMKTFM 694
>TC220076 homologue to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (39%)
Length = 808
Score = 240 bits (612), Expect = 2e-63
Identities = 117/227 (51%), Positives = 148/227 (64%), Gaps = 1/227 (0%)
Frame = +2
Query: 378 AASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRE 437
A G GF+A+DI F N AG KHQAVA R GSD +V +RC+ G+QD Y HSNRQF+R+
Sbjct: 80 AVKGKGFIAKDIGFVNNAGASKHQAVAFRSGSDRSVFFRCSFNGFQDTLYAHSNRQFYRD 259
Query: 438 CNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPA 497
C+I GT+DFIFGNAA VFQ C I R+P+ Q NTITAQ +KD NQNTGI I + P
Sbjct: 260 CDITGTIDFIFGNAAAVFQNCKIMPRQPLPNQFNTITAQGKKDRNQNTGIIIQKSKFTPL 439
Query: 498 PDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDF-ALKTLYYGEYMN 556
+ ++ TYLGRPWK +S TV M S +G + P GW+ W + + T++Y EY N
Sbjct: 440 ENNLTA----PTYLGRPWKDFSTTVIMQSDIGSFLKPVGWMSWVPNVEPVSTIFYAEYQN 607
Query: 557 FGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAF 603
GPGA + QRVKW GY+ + EA ++TV FI G WLP+ V F
Sbjct: 608 TGPGADVSQRVKWAGYKPTLTDGEAGKFTVQSFIQGPEWLPNAAVQF 748
>TC218157 similar to UP|Q8GUF8 (Q8GUF8) At2g47550/T30B22.15, partial (49%)
Length = 748
Score = 240 bits (612), Expect = 2e-63
Identities = 115/187 (61%), Positives = 134/187 (71%)
Frame = +1
Query: 421 GYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKD 480
GYQD YVHS RQF+ EC GTVDFIFGNA VVFQ CN+Y R PM+ Q N ITAQ R D
Sbjct: 4 GYQDTLYVHSLRQFYSECXXXGTVDFIFGNAKVVFQNCNMYPRLPMSGQFNAITAQGRTD 183
Query: 481 PNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEW 540
PNQ+TGISIH+ I A DLASS G + TYLGRPWK YSRTVYM ++M +H GW EW
Sbjct: 184 PNQDTGISIHNSTIRAADDLASSNG-VATYLGRPWKEYSRTVYMQTFMDSVIHAKGWREW 360
Query: 541 NGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTG 600
+GDFAL TLYY EY N GPG+ RV WPGY VI +T +A+ +TV+ F+ G WLP TG
Sbjct: 361 DGDFALSTLYYAEYSNSGPGSGTDNRVTWPGYHVINAT-DASNFTVSNFLLGDDWLPQTG 537
Query: 601 VAFLAGL 607
V++ L
Sbjct: 538 VSYTNNL 558
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,096,333
Number of Sequences: 63676
Number of extensions: 343485
Number of successful extensions: 2275
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 2158
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2184
length of query: 609
length of database: 12,639,632
effective HSP length: 103
effective length of query: 506
effective length of database: 6,081,004
effective search space: 3076988024
effective search space used: 3076988024
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC134822.9


