

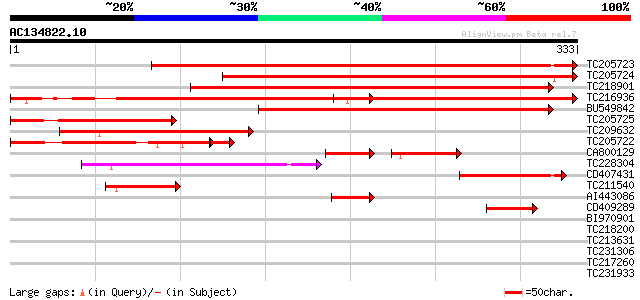
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.10 - phase: 0
(333 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205723 homologue to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), ... 520 e-148
TC205724 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), pa... 399 e-111
TC218901 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), pa... 389 e-108
TC216936 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), pa... 320 4e-88
BU549842 289 1e-78
TC205725 homologue to UP|Q9SPH3 (Q9SPH3) SINAH1 protein, partial... 164 4e-41
TC209632 similar to UP|Q9XGC3 (Q9XGC3) SINA2p, partial (34%) 148 4e-36
TC205722 homologue to UP|Q9SPH3 (Q9SPH3) SINAH1 protein, partial... 125 1e-32
CA800129 78 7e-29
TC228304 weakly similar to UP|Q9FKD3 (Q9FKD3) Similarity to S-lo... 103 1e-22
CD407431 similar to GP|21536945|gb| seven in absentia-like prote... 84 1e-16
TC211540 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), pa... 75 3e-14
AI443086 homologue to GP|21536945|gb| seven in absentia-like pro... 57 2e-08
CD409289 50 1e-06
BI970901 40 0.002
TC218200 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partia... 37 0.016
TC213631 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 35 0.047
TC231306 similar to UP|Q9LMD9 (Q9LMD9) F14D16.3, partial (18%) 35 0.062
TC217260 35 0.062
TC231933 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 34 0.11
>TC205723 homologue to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial
(65%)
Length = 1165
Score = 520 bits (1338), Expect = e-148
Identities = 239/250 (95%), Positives = 245/250 (97%)
Frame = +2
Query: 84 QCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYY 143
+CHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYY
Sbjct: 215 KCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYY 394
Query: 144 SKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPR 203
SKLKHET+CNFRPY+CPYAGSECS VGDI FLVAHLRDDHKVDMHTGCTFNHRYVKSNPR
Sbjct: 395 SKLKHETVCNFRPYSCPYAGSECSVVGDIPFLVAHLRDDHKVDMHTGCTFNHRYVKSNPR 574
Query: 204 EVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANG 263
EVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNY+YSLEVGANG
Sbjct: 575 EVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYSYSLEVGANG 754
Query: 264 RKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPD 323
RKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKE QN D
Sbjct: 755 RKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKE-QNSD 931
Query: 324 GGVCIPNLCS 333
GVC+PNLCS
Sbjct: 932 AGVCMPNLCS 961
>TC205724 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial (54%)
Length = 769
Score = 399 bits (1025), Expect = e-111
Identities = 186/210 (88%), Positives = 192/210 (90%), Gaps = 2/210 (0%)
Frame = +1
Query: 126 ELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKV 185
E CK S CPEIFPYYS KH +CNFRPY+C YAGSECS VGDI FLVAHLRDDHKV
Sbjct: 1 EXXCKXXSXXCPEIFPYYSXXKHXXVCNFRPYSCXYAGSECSVVGDIPFLVAHLRDDHKV 180
Query: 186 DMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMG 245
DMH GCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMG
Sbjct: 181 DMHPGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMG 360
Query: 246 DENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRK 305
DENEARNY+YSLEVGANGRKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFSGGDRK
Sbjct: 361 DENEARNYSYSLEVGANGRKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFSGGDRK 540
Query: 306 ELKLRVTGRIWKE--QQNPDGGVCIPNLCS 333
ELKLRVTGRIWKE QQN + GVC+PNLCS
Sbjct: 541 ELKLRVTGRIWKEQQQQNSEAGVCMPNLCS 630
>TC218901 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial (56%)
Length = 1058
Score = 389 bits (999), Expect = e-108
Identities = 181/213 (84%), Positives = 194/213 (90%)
Frame = +1
Query: 107 RQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSEC 166
R ELG+IRCLALEKVA SLELPCKY GC I+PYYSKLKHE+ C RPY CPYAGSEC
Sbjct: 1 RHELGNIRCLALEKVAASLELPCKYQGFGCIGIYPYYSKLKHESQCAHRPYNCPYAGSEC 180
Query: 167 SAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTVFHCFGQYFCLHF 226
S +GDI +LVAHL+DDHKVDMH G TFNHRYVKSNP+EVENATWMLTVF CFGQYFCLHF
Sbjct: 181 SIMGDIPYLVAHLKDDHKVDMHNGSTFNHRYVKSNPQEVENATWMLTVFSCFGQYFCLHF 360
Query: 227 EAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSH 286
EAFQLGMAPVY+AFLRFMGD+NEA+NY+YSLEVG NGRK+IW+G PRSIRDSHRKVRDS
Sbjct: 361 EAFQLGMAPVYIAFLRFMGDDNEAKNYSYSLEVGGNGRKMIWQGVPRSIRDSHRKVRDSF 540
Query: 287 DGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
DGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ
Sbjct: 541 DGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 639
>TC216936 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial (70%)
Length = 1648
Score = 320 bits (821), Expect = 4e-88
Identities = 159/217 (73%), Positives = 169/217 (77%), Gaps = 3/217 (1%)
Frame = +1
Query: 1 MDLDSIEC--VSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSL-KPRSGGNNNHGVIGST 57
M+ SIE VSS M+EDE HPH +FSS+ K S G
Sbjct: 319 MESSSIESSTVSSMTMMEEDE------HPH--------QFSSISKLHSNGPTT------- 435
Query: 58 AIAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLA 117
TSVH+LLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLA
Sbjct: 436 -----TSVHDLLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLA 600
Query: 118 LEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGSECSAVGDINFLVA 177
LEK+AESLELPC+Y SLGCPEIFPYYSKLKHE ICNFRPY CPYAGS+CS VGDI LVA
Sbjct: 601 LEKIAESLELPCRYISLGCPEIFPYYSKLKHEAICNFRPYNCPYAGSDCSVVGDIPCLVA 780
Query: 178 HLRDDHKVDMHTGCTFNHRYVKSNPREVENATWMLTV 214
HLRDDH+VDMH+GCTFNHRYVKSNP EVENATWMLTV
Sbjct: 781 HLRDDHRVDMHSGCTFNHRYVKSNPMEVENATWMLTV 891
Score = 239 bits (611), Expect = 1e-63
Identities = 115/145 (79%), Positives = 126/145 (86%), Gaps = 2/145 (1%)
Frame = +2
Query: 191 CTFNHRY--VKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDEN 248
CT H + + P ++ + + VFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDE
Sbjct: 995 CTSTHLHFGLLQIPSKLLTHSCLC*VFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDER 1174
Query: 249 EARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELK 308
EARNY+YSLEVG NGRKL +EG+PRSIRDSH+KVRDSHDGLII RNMALFFSGGDRKELK
Sbjct: 1175 EARNYSYSLEVGGNGRKLTFEGSPRSIRDSHKKVRDSHDGLIIYRNMALFFSGGDRKELK 1354
Query: 309 LRVTGRIWKEQQNPDGGVCIPNLCS 333
LRVTGRIWKEQQNP+GGVCIPNLCS
Sbjct: 1355 LRVTGRIWKEQQNPEGGVCIPNLCS 1429
>BU549842
Length = 710
Score = 289 bits (739), Expect = 1e-78
Identities = 132/173 (76%), Positives = 150/173 (86%)
Frame = -2
Query: 147 KHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPREVE 206
KHE C RPY C A E +GDI VAHL+DDHKVDMH GCTFNHRYVK+NP EVE
Sbjct: 709 KHEQNCGXRPYNCXXAXXEXXVMGDIXTXVAHLKDDHKVDMHDGCTFNHRYVKANPHEVE 530
Query: 207 NATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRKL 266
NATWMLTVF+ FG++FCLH EAFQLG APVYMAFLRFMGD+NEA+ ++YSLEVGANGRKL
Sbjct: 529 NATWMLTVFNSFGRHFCLHXEAFQLGSAPVYMAFLRFMGDDNEAKKFSYSLEVGANGRKL 350
Query: 267 IWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
IW+G PRSIRDSHRKVRDS DGLIIQRN+AL+FSGG+R+ELKLR+TGRIW+E+
Sbjct: 349 IWQGIPRSIRDSHRKVRDSQDGLIIQRNLALYFSGGERQELKLRITGRIWREE 191
>TC205725 homologue to UP|Q9SPH3 (Q9SPH3) SINAH1 protein, partial (14%)
Length = 662
Score = 164 bits (416), Expect = 4e-41
Identities = 76/98 (77%), Positives = 81/98 (82%)
Frame = +3
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
MDL+S+ECVSSSDGMDEDEI H HHHHSEFSS KPR+GG +N +G AIA
Sbjct: 393 MDLESVECVSSSDGMDEDEI--------HANHHHHSEFSSTKPRNGGTSNINSVGPNAIA 548
Query: 61 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR
Sbjct: 549 PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 662
>TC209632 similar to UP|Q9XGC3 (Q9XGC3) SINA2p, partial (34%)
Length = 558
Score = 148 bits (373), Expect = 4e-36
Identities = 69/121 (57%), Positives = 83/121 (68%), Gaps = 7/121 (5%)
Frame = +3
Query: 30 QQHHHHSEFSSLKPRSGGNNNH-------GVIGSTAIAPATSVHELLECPVCTNSMYPPI 82
+ H S++ K +S +N G+ G + I+ V+ELL CPVC N MYPPI
Sbjct: 195 ESHLMSSDYEMGKAKSEAKSNSTSTKSSIGLSGKSGISSNNGVYELLGCPVCKNLMYPPI 374
Query: 83 HQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPY 142
HQC NGHTLCS CK VHN CP+C +LG+IRCLALEKVAESLELPC+Y SLGC +IFPY
Sbjct: 375 HQCPNGHTLCSHCKVEVHNICPSCHHDLGNIRCLALEKVAESLELPCRYQSLGCHDIFPY 554
Query: 143 Y 143
Y
Sbjct: 555 Y 557
>TC205722 homologue to UP|Q9SPH3 (Q9SPH3) SINAH1 protein, partial (24%)
Length = 728
Score = 125 bits (315), Expect(2) = 1e-32
Identities = 71/127 (55%), Positives = 80/127 (62%), Gaps = 7/127 (5%)
Frame = +3
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIA 60
MDL S+ECVSSSDGMDEDEI +HHHSEFSS KPR+GG +N +G IA
Sbjct: 342 MDLMSVECVSSSDGMDEDEIH---------ANHHHSEFSSTKPRNGGTSNINSVGPNGIA 494
Query: 61 PATSVHELLECPVCTNSMYPPIHQC---HNGHTLCSTCKTRVH----NRCPTCRQELGDI 113
PATSVHELLECPVC + QC T+ + C V RCPTCRQELGDI
Sbjct: 495 PATSVHELLECPVC-------LIQCTLQFISVTMATHCVPHVKLGCTTRCPTCRQELGDI 653
Query: 114 RCLALEK 120
CLALE+
Sbjct: 654 XCLALEE 674
Score = 32.0 bits (71), Expect(2) = 1e-32
Identities = 13/14 (92%), Positives = 14/14 (99%)
Frame = +1
Query: 119 EKVAESLELPCKYY 132
+KVAESLELPCKYY
Sbjct: 670 KKVAESLELPCKYY 711
>CA800129
Length = 199
Score = 78.2 bits (191), Expect(2) = 7e-29
Identities = 38/43 (88%), Positives = 40/43 (92%), Gaps = 2/43 (4%)
Frame = +3
Query: 225 HFEA--FQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANGRK 265
H +A FQLGMAPVYMAFLRFMGDENEARNY+YSLEVGANGRK
Sbjct: 69 HLDAHTFQLGMAPVYMAFLRFMGDENEARNYSYSLEVGANGRK 197
Score = 67.0 bits (162), Expect(2) = 7e-29
Identities = 28/29 (96%), Positives = 29/29 (99%)
Frame = +1
Query: 186 DMHTGCTFNHRYVKSNPREVENATWMLTV 214
DMHTGCTFNHRYVKSNPREVENATWMLT+
Sbjct: 1 DMHTGCTFNHRYVKSNPREVENATWMLTL 87
>TC228304 weakly similar to UP|Q9FKD3 (Q9FKD3) Similarity to S-locus protein,
partial (33%)
Length = 1129
Score = 103 bits (256), Expect = 1e-22
Identities = 51/145 (35%), Positives = 72/145 (49%), Gaps = 4/145 (2%)
Frame = +1
Query: 43 PRSGGNNNHGVIGSTA----IAPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTR 98
P G N+N G+++ + S ++L+C +C + P+ QC NGH CSTC R
Sbjct: 175 PHDGSNSNANGAGTSSRDRSVPIFVSDPDVLDCCICYEPLAAPVFQCENGHIACSTCCVR 354
Query: 99 VHNRCPTCRQELGDIRCLALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYT 158
+ N+CP C +G RC A+EKV E +++ C + GC E F Y K HE C + P +
Sbjct: 355 LSNKCPMCLMPIGYNRCRAIEKVLECIKMSCPNANYGCKETFSYSRKNNHEKECIYLPCS 534
Query: 159 CPYAGSECSAVGDINFLVAHLRDDH 183
CP G C V L H H
Sbjct: 535 CPLTG--CDFVASSKELFLHFSHRH 603
>CD407431 similar to GP|21536945|gb| seven in absentia-like protein
{Arabidopsis thaliana}, partial (11%)
Length = 289
Score = 83.6 bits (205), Expect = 1e-16
Identities = 45/66 (68%), Positives = 53/66 (80%), Gaps = 3/66 (4%)
Frame = -3
Query: 265 KLIWEGTPRSI-RDSHRKVR-DSHDGLIIQRN-MALFFSGGDRKELKLRVTGRIWKEQQN 321
KLIWEGTPR + RDSHRKV+ +SHDGLI+ + MALFFSGGDRK + + +GRIWKE QN
Sbjct: 287 KLIWEGTPREVFRDSHRKVKGNSHDGLILFKEIMALFFSGGDRKGAEAQESGRIWKE-QN 111
Query: 322 PDGGVC 327
D GVC
Sbjct: 110 SDAGVC 93
>TC211540 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial (10%)
Length = 497
Score = 75.5 bits (184), Expect = 3e-14
Identities = 33/48 (68%), Positives = 38/48 (78%), Gaps = 4/48 (8%)
Frame = +3
Query: 57 TAIAP----ATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVH 100
TA+ P ++SV ELLECPVC N+MYPPIHQC NGHT+CS CK RVH
Sbjct: 354 TALKPNGTVSSSVRELLECPVCLNAMYPPIHQCSNGHTICSGCKPRVH 497
>AI443086 homologue to GP|21536945|gb| seven in absentia-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 322
Score = 56.6 bits (135), Expect = 2e-08
Identities = 24/25 (96%), Positives = 24/25 (96%)
Frame = +3
Query: 190 GCTFNHRYVKSNPREVENATWMLTV 214
GCTFNHRYVKSNP EVENATWMLTV
Sbjct: 3 GCTFNHRYVKSNPMEVENATWMLTV 77
Score = 39.7 bits (91), Expect = 0.002
Identities = 31/88 (35%), Positives = 39/88 (44%), Gaps = 9/88 (10%)
Frame = +2
Query: 154 FRPYTCPYAGSECSAVG------DINFLVAHLRDDHKVDMHTGCTFNHRYV---KSNPRE 204
+ YT P G CSAV D+ F+ HL+D H Y+ KS+P
Sbjct: 80 YNNYTRPSLG--CSAVHGMHLIVDMTFIAMHLQDIPSFHAQA-----HTYILDFKSHPNC 238
Query: 205 VENATWMLTVFHCFGQYFCLHFEAFQLG 232
++ F F FCLHFEAFQLG
Sbjct: 239 YG*LIFVYVSFPLFWSDFCLHFEAFQLG 322
>CD409289
Length = 332
Score = 50.4 bits (119), Expect = 1e-06
Identities = 23/30 (76%), Positives = 27/30 (89%)
Frame = -1
Query: 281 KVRDSHDGLIIQRNMALFFSGGDRKELKLR 310
KVRDS DGLIIQRNMALFFSG DRK+++ +
Sbjct: 299 KVRDSFDGLIIQRNMALFFSGDDRKDIEAK 210
Score = 31.2 bits (69), Expect = 0.68
Identities = 15/22 (68%), Positives = 16/22 (72%)
Frame = -2
Query: 298 FFSGGDRKELKLRVTGRIWKEQ 319
FF + LKLRVTGRIWKEQ
Sbjct: 247 FFPVMTGRILKLRVTGRIWKEQ 182
>BI970901
Length = 447
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/55 (41%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Frame = -3
Query: 272 PRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKL-RVTGRIWKEQQNPDGG 325
PRSIRD +KV SHDGLI N+ F D E + K+ QN +GG
Sbjct: 376 PRSIRDXLKKVTGSHDGLITVHNIERFCLQADTVEKSICEYQAEYCKDHQNREGG 212
Score = 36.2 bits (82), Expect = 0.021
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 4/71 (5%)
Frame = -2
Query: 249 EARNYTYSLEVGANGRKLIWEGTPRS----IRDSHRKVRDSHDGLIIQRNMALFFSGGDR 304
EAR + S EVG N KL +G+ + + HR+ SH ++ AL F+GG
Sbjct: 446 EARILSSSWEVGGNXXKLS*DGSAKEH*R*X*EGHRQS*WSHHS---PQHRALLFAGGYS 276
Query: 305 KELKLRVTGRI 315
+E+ +RV GR+
Sbjct: 275 REINMRVPGRV 243
>TC218200 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partial (25%)
Length = 1175
Score = 36.6 bits (83), Expect = 0.016
Identities = 29/116 (25%), Positives = 49/116 (42%), Gaps = 14/116 (12%)
Frame = +1
Query: 63 TSVHELLE------CPVCTNSMYPPIHQC------HNGHTLCSTCKTRVHNRCPTCRQEL 110
+S H+ LE CP+C + ++ H H+ C T H CP C + L
Sbjct: 601 SSSHKCLEKGLEMNCPICCDDLFTSSATVRALPCGHYMHSACFQAYTCSHYTCPICSKSL 780
Query: 111 GDIRCL--ALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGS 164
GD+ L+ + + ELP + Y C +I + K + ++ + C + GS
Sbjct: 781 GDMAVYFGMLDALLAAEELP-EEYKDRCQDILCHDCDRKGTSRFHWLYHKCGFCGS 945
>TC213631 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(32%)
Length = 589
Score = 35.0 bits (79), Expect = 0.047
Identities = 24/82 (29%), Positives = 35/82 (42%), Gaps = 6/82 (7%)
Frame = +3
Query: 60 APATSVHELLECPVCTNSMYPP-----IHQCHNG-HTLCSTCKTRVHNRCPTCRQELGDI 113
A A S + +C +C + QC +G H C R H+ CP+CRQ L
Sbjct: 39 ATAESAVKFADCAICLTEFAAGDEIRVLPQCGHGFHVSCIDAWLRSHSSCPSCRQILVVS 218
Query: 114 RCLALEKVAESLELPCKYYSLG 135
RC + + S EL + + G
Sbjct: 219 RCDKVRRDPGSGELQLRSAAAG 284
>TC231306 similar to UP|Q9LMD9 (Q9LMD9) F14D16.3, partial (18%)
Length = 778
Score = 34.7 bits (78), Expect = 0.062
Identities = 29/108 (26%), Positives = 49/108 (44%), Gaps = 14/108 (12%)
Frame = +2
Query: 71 CPVCTNSMYP---PIHQCHNGHTLCSTC---KTRVHNRCPTCRQELGDIR--------CL 116
CP+C ++ P+ GH + STC T + CP C + LGD++ L
Sbjct: 407 CPICHEYIFTSCSPVKALPCGHVMHSTCFQEYTCFNYTCPICSKSLGDMQVYFRMLDALL 586
Query: 117 ALEKVAESLELPCKYYSLGCPEIFPYYSKLKHETICNFRPYTCPYAGS 164
A E++++ E+ + L C + + K ET ++ + CP GS
Sbjct: 587 AEERISD--EISSQTQVLLCND-----CEKKGETPFHWLYHKCPSCGS 709
>TC217260
Length = 1114
Score = 34.7 bits (78), Expect = 0.062
Identities = 18/61 (29%), Positives = 26/61 (42%), Gaps = 5/61 (8%)
Frame = +3
Query: 55 GSTAIAPATSVHELLECPVCTNSMYPPIHQCHN--GHTLCSTC---KTRVHNRCPTCRQE 109
GS P + CP+C M P +H+ GH C C +CPTCR++
Sbjct: 459 GSVKKTPEPPKELVFNCPIC---MGPMVHEMSTRCGHIFCKDCIKAAISAQGKCPTCRKK 629
Query: 110 L 110
+
Sbjct: 630 V 632
>TC231933 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(65%)
Length = 619
Score = 33.9 bits (76), Expect = 0.11
Identities = 20/62 (32%), Positives = 27/62 (43%), Gaps = 6/62 (9%)
Frame = +2
Query: 60 APATSVHELLECPVCTNSMYPP-----IHQCHNG-HTLCSTCKTRVHNRCPTCRQELGDI 113
A A S + +C +C + QC +G H C R H+ CP+CRQ L
Sbjct: 278 ASAESAVKFADCAICLTEFAAGDEIRVLPQCGHGFHVSCIDAWLRSHSSCPSCRQILVVS 457
Query: 114 RC 115
RC
Sbjct: 458 RC 463
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,794,194
Number of Sequences: 63676
Number of extensions: 411971
Number of successful extensions: 4934
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 3796
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4493
length of query: 333
length of database: 12,639,632
effective HSP length: 98
effective length of query: 235
effective length of database: 6,399,384
effective search space: 1503855240
effective search space used: 1503855240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC134822.10


