

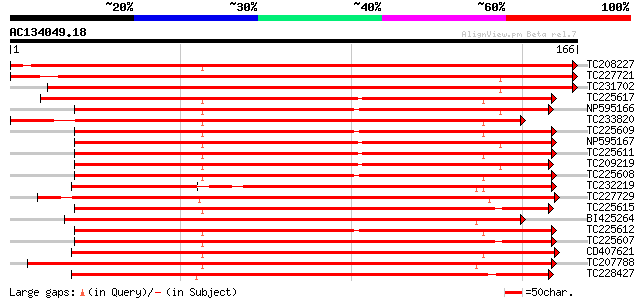
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.18 - phase: 0
(166 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208227 weakly similar to UP|Q9ZS32 (Q9ZS32) NL25, partial (17%) 250 3e-67
TC227721 similar to UP|Q8H0N7 (Q8H0N7) TIR-NBS disease resistanc... 225 7e-60
TC231702 weakly similar to UP|Q947E1 (Q947E1) Resistance gene an... 194 2e-50
TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete 145 9e-36
NP595166 R 14 protein [Glycine max] 141 2e-34
TC233820 similar to UP|Q84ZV3 (Q84ZV3) R 4 protein, partial (15%) 141 2e-34
TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete 138 1e-33
NP595167 R 13 protein [Glycine max] 137 2e-33
TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete 136 4e-33
TC209219 UP|Q9FVK2 (Q9FVK2) Resistance protein MG13 (Fragment), ... 135 7e-33
TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete 135 9e-33
TC232219 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain... 134 2e-32
TC227729 134 3e-32
TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete 134 3e-32
BI425264 weakly similar to GP|28558777|g MRGH5 {Cucumis melo}, p... 133 3e-32
TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete 132 6e-32
TC225607 homologue to UP|Q84ZV1 (Q84ZV1) R 9 protein, complete 132 1e-31
CD407621 132 1e-31
TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate re... 129 5e-31
TC228427 UP|Q8H6S7 (Q8H6S7) Resistance protein KR3, complete 129 5e-31
>TC208227 weakly similar to UP|Q9ZS32 (Q9ZS32) NL25, partial (17%)
Length = 697
Score = 250 bits (638), Expect = 3e-67
Identities = 123/167 (73%), Positives = 147/167 (87%), Gaps = 1/167 (0%)
Frame = +1
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSG 59
MAWS SS+SS++NTP +KHEVFISFRSEDTR FTSHLN AL+RLDI+TY+DNN L+ G
Sbjct: 52 MAWS--SSSSSTSNTPPQKHEVFISFRSEDTRKTFTSHLNAALERLDIKTYLDNNNLDRG 225
Query: 60 DEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTD 119
+EI TTLVRAIEEA+LSVIVFSKNYA SK+CLDEL+KILE R K +++P+FYD+DP+D
Sbjct: 226 EEIPTTLVRAIEEAKLSVIVFSKNYADSKWCLDELLKILEFGRAKTLIIMPVFYDIDPSD 405
Query: 120 VRNQRGSYAEAFAKHEKNSEEKIKVQEWRNGLMEAANYSGWDCNVNR 166
VRNQRG+YAEAF KHE+ +EK K+QEWR GL+EAANYSGWDC+VNR
Sbjct: 406 VRNQRGTYAEAFDKHERYFQEKKKLQEWRKGLVEAANYSGWDCDVNR 546
>TC227721 similar to UP|Q8H0N7 (Q8H0N7) TIR-NBS disease resistance protein,
partial (19%)
Length = 1015
Score = 225 bits (574), Expect = 7e-60
Identities = 110/167 (65%), Positives = 137/167 (81%), Gaps = 1/167 (0%)
Frame = +2
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGD 60
MAWS SSS+ TP +KHEVF+SFR EDTR FT HL+ +L RL + TYID NL G+
Sbjct: 56 MAWSTSSSS-----TPHQKHEVFLSFRGEDTRYTFTGHLHASLTRLQVNTYIDYNLQRGE 220
Query: 61 EISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDV 120
EIS++L+RAIEEA+LSV+VFSKNY SK+CLDEL+KILECK M+G++V+PIFYD+DP+ V
Sbjct: 221 EISSSLLRAIEEAKLSVVVFSKNYGNSKWCLDELLKILECKNMRGQIVLPIFYDIDPSHV 400
Query: 121 RNQRGSYAEAFAKHEKNSEEKI-KVQEWRNGLMEAANYSGWDCNVNR 166
RNQ G+YAEAFAKHEK+ + ++ KVQ+WR L EAAN SGWDC+VNR
Sbjct: 401 RNQTGTYAEAFAKHEKHLQGQMDKVQKWRVALREAANLSGWDCSVNR 541
>TC231702 weakly similar to UP|Q947E1 (Q947E1) Resistance gene analog NBS7
(Fragment), partial (40%)
Length = 935
Score = 194 bits (492), Expect = 2e-50
Identities = 93/156 (59%), Positives = 120/156 (76%), Gaps = 1/156 (0%)
Frame = +1
Query: 12 SNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIE 71
++ TPQ+ H+VF+SFR EDTR FTSHL AL RL ++TYIDN L GDEIS +L+RAI+
Sbjct: 13 TSKTPQQVHDVFLSFRGEDTRYTFTSHLYAALTRLQVKTYIDNELERGDEISPSLLRAID 192
Query: 72 EAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAF 131
+A+++VIVFS+NYA+S++CLDEL+KI+ECKR G+++VP+FY VDPT VR+Q GSY AF
Sbjct: 193 DAKVAVIVFSENYASSRWCLDELVKIMECKRKNGQIIVPVFYHVDPTHVRHQTGSYGHAF 372
Query: 132 AKHEKNSEEKI-KVQEWRNGLMEAANYSGWDCNVNR 166
A HE+ + KVQ WR L E AN SGWDC R
Sbjct: 373 AMHEQRFVGNMNKVQTWRLVLGEVANISGWDCLTTR 480
>TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete
Length = 2697
Score = 145 bits (366), Expect = 9e-36
Identities = 74/153 (48%), Positives = 109/153 (70%), Gaps = 2/153 (1%)
Frame = +1
Query: 10 SSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVR 68
+++ +P ++VF+SFR DTR+ FT +L AL I T+ID+ L GDEI+ L +
Sbjct: 4 AATTRSPASIYDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSK 183
Query: 69 AIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYA 128
AI+E+ +++ V S+NYA+S FCLDEL+ +L CKR KG +V+P+FY+VDP+DVR Q+GSY
Sbjct: 184 AIQESRIAITVLSQNYASSSFCLDELVTVLLCKR-KGLLVIPVFYNVDPSDVRQQKGSYG 360
Query: 129 EAFAKHEKN-SEEKIKVQEWRNGLMEAANYSGW 160
EA AKH+K +K K+Q+WR L + A+ SG+
Sbjct: 361 EAMAKHQKRFKAKKEKLQKWRMALHQVADLSGY 459
>NP595166 R 14 protein [Glycine max]
Length = 1926
Score = 141 bits (355), Expect = 2e-34
Identities = 71/142 (50%), Positives = 101/142 (71%), Gaps = 2/142 (1%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF+SFR EDTR FT +L AL I T+ D L+ GDEI+ L +AI+E+ +++
Sbjct: 34 YDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSKAIQESRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNS 138
V S+NYA S FCLDEL+ IL CK +G +V+P+FY+VDP+D+R+Q+GSY EA KH+K
Sbjct: 214 VLSQNYAFSSFCLDELVTILHCK-SEGLLVIPVFYNVDPSDLRHQKGSYGEAMIKHQKRF 390
Query: 139 EEKI-KVQEWRNGLMEAANYSG 159
E K+ K+Q+WR L + A+ SG
Sbjct: 391 ESKMEKLQKWRMALKQVADLSG 456
>TC233820 similar to UP|Q84ZV3 (Q84ZV3) R 4 protein, partial (15%)
Length = 471
Score = 141 bits (355), Expect = 2e-34
Identities = 72/153 (47%), Positives = 104/153 (67%), Gaps = 2/153 (1%)
Frame = +1
Query: 1 MAWSYSSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSG 59
MAW SS+SSSN ++VF+SFR EDTRN FT HL L+ I T+ID+ L G
Sbjct: 28 MAWGSRSSSSSSN------YDVFLSFRGEDTRNAFTGHLYNTLQSKGIHTFIDDEKLQRG 189
Query: 60 DEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTD 119
++I+ L++AI+++ +++ V S++YA+S FCLDEL IL C + K +V+P+FY VDP+D
Sbjct: 190 EQITPALMKAIQDSRVAITVLSEHYASSSFCLDELATILHCDQRKRLLVIPVFYKVDPSD 369
Query: 120 VRNQRGSYAEAFAKHEKN-SEEKIKVQEWRNGL 151
VR+Q+GSY EA AK E+ + K+Q W+ L
Sbjct: 370 VRHQKGSYGEALAKLERRFQHDPEKLQNWKMAL 468
>TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete
Length = 2688
Score = 138 bits (348), Expect = 1e-33
Identities = 72/143 (50%), Positives = 102/143 (70%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF+SFR DTR+ FT +L AL I T ID+ L GDEI+ L +AI+E+ +++
Sbjct: 34 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN- 137
V S+NYA+S FCLDEL+ IL CK +G +V+P+FY VDP+DVR+Q+GSY EA AKH+K
Sbjct: 214 VLSQNYASSSFCLDELVTILHCK-SEGLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 390
Query: 138 SEEKIKVQEWRNGLMEAANYSGW 160
+K K+Q+WR L + A+ SG+
Sbjct: 391 KAKKEKLQKWRMALKQVADLSGY 459
>NP595167 R 13 protein [Glycine max]
Length = 1926
Score = 137 bits (346), Expect = 2e-33
Identities = 71/143 (49%), Positives = 99/143 (68%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF+SFR DTRN FT +L AL I T+ID+ L GD+I+ L AI E+ +++
Sbjct: 34 YDVFLSFRGLDTRNGFTGNLYKALGDRGIYTFIDDQELPRGDKITPALSNAINESRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNS 138
V S+NYA S FCLDEL+ IL CK +G +V+P+FY VDP+DVR+Q+GSY E KH+K
Sbjct: 214 VLSENYAFSSFCLDELVTILHCKS-EGLLVIPVFYKVDPSDVRHQKGSYGETMTKHQKRF 390
Query: 139 EEKI-KVQEWRNGLMEAANYSGW 160
E K+ K++EWR L + A+ SG+
Sbjct: 391 ESKMEKLREWRMALQQVADLSGY 459
>TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete
Length = 2709
Score = 136 bits (343), Expect = 4e-33
Identities = 69/143 (48%), Positives = 104/143 (72%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF++FR EDTR FT +L AL I T+ D + L+SGD+I+ L +AI+E+ +++
Sbjct: 34 YDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSKAIQESRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN- 137
V S+NYA+S FCLDEL+ IL CKR +G +V+P+F++VDP+ VR+ +GSY EA AKH+K
Sbjct: 214 VLSQNYASSSFCLDELVTILHCKR-EGLLVIPVFHNVDPSAVRHLKGSYGEAMAKHQKRF 390
Query: 138 SEEKIKVQEWRNGLMEAANYSGW 160
+K K+Q+WR L + A+ SG+
Sbjct: 391 KAKKEKLQKWRMALHQVADLSGY 459
>TC209219 UP|Q9FVK2 (Q9FVK2) Resistance protein MG13 (Fragment), complete
Length = 1224
Score = 135 bits (341), Expect = 7e-33
Identities = 68/142 (47%), Positives = 102/142 (70%), Gaps = 2/142 (1%)
Frame = +3
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF+SFR DTR FT +L AL T+ D + L+SG+EI+ L++AI+++ +++I
Sbjct: 63 YDVFLSFRGTDTRYGFTGNLYKALCDKGFHTFFDEDKLHSGEEITPALLKAIQDSRVAII 242
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNS 138
V S+NYA S FCLDEL+ I CKR +G +V+P+FY VDP+ VR+Q+GSY EA KH++
Sbjct: 243 VLSENYAFSSFCLDELVTIFHCKR-EGLLVIPVFYKVDPSYVRHQKGSYGEAMTKHQERF 419
Query: 139 EEKI-KVQEWRNGLMEAANYSG 159
++K+ K+QEWR L + A+ SG
Sbjct: 420 KDKMEKLQEWRMALKQVADLSG 485
>TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete
Length = 3084
Score = 135 bits (340), Expect = 9e-33
Identities = 70/143 (48%), Positives = 98/143 (67%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF+SF +DTR FT +L AL I T+ID+ L GDEI L AI+E+ +++
Sbjct: 34 YDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN- 137
V S+NYA+S FCLDEL+ IL CK +G +V+P+FY VDP+ VR+Q+GSY EA AKH+K
Sbjct: 214 VLSQNYASSSFCLDELVTILHCK-SQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRF 390
Query: 138 SEEKIKVQEWRNGLMEAANYSGW 160
K K+Q+WR L + A+ SG+
Sbjct: 391 KANKEKLQKWRMALHQVADLSGY 459
>TC232219 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (21%)
Length = 845
Score = 134 bits (337), Expect = 2e-32
Identities = 70/143 (48%), Positives = 96/143 (66%), Gaps = 1/143 (0%)
Frame = +2
Query: 19 KHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELSVI 78
+++VF+SF DTR +FT L AL R +TY++++ GD+IS + I ++ LS+I
Sbjct: 401 RNDVFLSFCGRDTRYSFTGFLYNALSRSGFKTYMNDD---GDQISQS---TIGKSRLSII 562
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN- 137
VFSKNYA S CLDEL+ ILEC +MK ++V PIFY V+P D+R QR SY EA +HE
Sbjct: 563 VFSKNYAHSSSCLDELLAILECMKMKNQLVWPIFYKVEPRDIRRQRNSYGEAMTEHENML 742
Query: 138 SEEKIKVQEWRNGLMEAANYSGW 160
++ KVQ+WR+ L EAA GW
Sbjct: 743 GKDSEKVQKWRSALFEAAXLXGW 811
Score = 85.5 bits (210), Expect = 1e-17
Identities = 39/106 (36%), Positives = 69/106 (64%), Gaps = 1/106 (0%)
Frame = +2
Query: 56 LNSGDEISTTLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDV 115
L GD+I+T ++ A+E + +S++VFS +A+S CLD+L+ I C K ++++PIFYDV
Sbjct: 5 LRRGDKIATAILTAMEASRISIVVFSPYFASSTCCLDQLVHIHRCMNTKNQLILPIFYDV 184
Query: 116 DPTDVRNQRGSYAEAFAKHE-KNSEEKIKVQEWRNGLMEAANYSGW 160
D +DVR+Q ++ +A +H+ + + KV +W + L AN + +
Sbjct: 185 DQSDVRDQLNTFGQAMLQHQHRFGKSSDKVLQWSSVLSHVANLTAF 322
>TC227729
Length = 1391
Score = 134 bits (336), Expect = 3e-32
Identities = 67/155 (43%), Positives = 108/155 (69%), Gaps = 2/155 (1%)
Frame = +2
Query: 9 TSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDN-NLNSGDEISTTLV 67
TSSS++ +++VF+SFR EDTRN+FT L ALK+ I + D+ ++ G+ I+ L+
Sbjct: 149 TSSSSSF---EYDVFVSFRGEDTRNSFTGFLFEALKKQGIEAFKDDKDIRKGESIAPELI 319
Query: 68 RAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSY 127
RAIE + + ++VFSK+YA+S +CL EL I C + ++++PIFYDVDP+ VR Q G Y
Sbjct: 320 RAIEGSHVFLVVFSKDYASSTWCLRELAHIWNCIQTSSRLLLPIFYDVDPSQVRKQSGDY 499
Query: 128 AEAFAKHEKNSE-EKIKVQEWRNGLMEAANYSGWD 161
+AF++H+++S ++ +++ WR L + A+ SGWD
Sbjct: 500 EKAFSQHQQSSRFQEKEIKTWREVLKQVASLSGWD 604
>TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete
Length = 2724
Score = 134 bits (336), Expect = 3e-32
Identities = 66/141 (46%), Positives = 98/141 (68%), Gaps = 1/141 (0%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF+SFR EDTR FT +L AL I T+ D + L+SG+EI+ L++AI+++ +++
Sbjct: 34 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNS 138
V S+++A+S FCLDEL IL C + G MV+P+FY V P DVR+Q+G+Y EA AKH+K
Sbjct: 214 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 393
Query: 139 EEKIKVQEWRNGLMEAANYSG 159
+K+ Q+W L + AN SG
Sbjct: 394 PDKL--QKWERALRQVANLSG 450
>BI425264 weakly similar to GP|28558777|g MRGH5 {Cucumis melo}, partial (9%)
Length = 421
Score = 133 bits (335), Expect = 3e-32
Identities = 59/136 (43%), Positives = 98/136 (71%), Gaps = 1/136 (0%)
Frame = +1
Query: 17 QEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNNLNSGDEISTTLVRAIEEAELS 76
+ +++VF+ FR EDTR FT +L AL++ +RT+ D+ SGD+I +++AI+E+ +S
Sbjct: 4 KRRYDVFLCFRGEDTRYTFTGNLYAALRQARLRTFFDDGFKSGDQIFDVVLQAIQESRIS 183
Query: 77 VIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHE- 135
++V S+N+A+S +CL+EL+KILEC+ K ++V+PIFY +DP+DVR Q G Y E+ A+H+
Sbjct: 184 IVVLSENFASSSWCLEELVKILECRETKKQLVIPIFYRMDPSDVRRQTGCYGESLAQHQY 363
Query: 136 KNSEEKIKVQEWRNGL 151
+ + KV+ W+ L
Sbjct: 364 EFRSDSEKVRNWQEAL 411
>TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete
Length = 2706
Score = 132 bits (333), Expect = 6e-32
Identities = 67/143 (46%), Positives = 100/143 (69%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF++FR DTR FT +L AL I T+ D L+ G+EI+ L++AI+E+ +++
Sbjct: 34 YDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLKAIQESRIAIT 213
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN- 137
V SKNYA+S FCLDEL+ IL CK +G +V+P+FY+VDP+DVR+Q+GSY AKH+K
Sbjct: 214 VLSKNYASSSFCLDELVTILHCK-SEGLLVIPVFYNVDPSDVRHQKGSYGVEMAKHQKRF 390
Query: 138 SEEKIKVQEWRNGLMEAANYSGW 160
+K K+Q+WR L + A+ G+
Sbjct: 391 KAKKEKLQKWRIALKQVADLCGY 459
>TC225607 homologue to UP|Q84ZV1 (Q84ZV1) R 9 protein, complete
Length = 1063
Score = 132 bits (331), Expect = 1e-31
Identities = 64/142 (45%), Positives = 98/142 (68%), Gaps = 1/142 (0%)
Frame = +3
Query: 20 HEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSVI 78
++VF++FR EDTR FTS+L AL IRT+ D L+SG+EI+ L++AI+++ +++
Sbjct: 60 YDVFLNFRGEDTRYGFTSNLYRALSDKGIRTFFDEEKLHSGEEITPALLKAIKDSRIAIT 239
Query: 79 VFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKNS 138
V S+++A+S FCLDEL I+ C + G M++P+FY V P+DVR+Q+G+Y EA AKH+
Sbjct: 240 VLSEDFASSSFCLDELTSIVHCAQYNGMMIIPVFYKVYPSDVRHQKGTYGEALAKHKIRF 419
Query: 139 EEKIKVQEWRNGLMEAANYSGW 160
EK Q W L + A+ SG+
Sbjct: 420 PEKF--QNWEMALRQVADLSGF 479
>CD407621
Length = 571
Score = 132 bits (331), Expect = 1e-31
Identities = 65/145 (44%), Positives = 91/145 (61%), Gaps = 2/145 (1%)
Frame = -3
Query: 19 KHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEISTTLVRAIEEAELSV 77
+++VFISFR DTRN F HL L R I + D+ L G+ IS L++AI+++ LS+
Sbjct: 569 RYDVFISFRGPDTRNTFVDHLYAHLLRKGIFVFKDDEKLQKGESISAQLLQAIQDSRLSI 390
Query: 78 IVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN 137
IVFSK YA+S +CLDE+ I +CK+ V P+FYDVDP+ VR+Q G+Y AF H
Sbjct: 389 IVFSKQYASSTWCLDEMAAIADCKQQSNPTVFPVFYDVDPSHVRHQNGAYEVAFVSHRSR 210
Query: 138 -SEEKIKVQEWRNGLMEAANYSGWD 161
E+ KV W + + A+ +GWD
Sbjct: 209 FREDPDKVDRWARAMTDLAHSAGWD 135
>TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate resistance
protein KR1, complete
Length = 3949
Score = 129 bits (325), Expect = 5e-31
Identities = 67/159 (42%), Positives = 107/159 (67%), Gaps = 4/159 (2%)
Frame = +1
Query: 6 SSSTSSSNNTPQEKHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYIDNN-LNSGDEIST 64
+ +SSS+ + + ++VF+SFR EDTR FT +L AL I T++D+ + GD+I++
Sbjct: 103 AKQSSSSSFSYRFSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQITS 282
Query: 65 TLVRAIEEAELSVIVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQR 124
L +AIEE+ + +IV S+NYA+S FCL+EL IL+ + KG +++P+FY VDP+DVRN
Sbjct: 283 GLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNHT 462
Query: 125 GSYAEAFAKHE---KNSEEKIKVQEWRNGLMEAANYSGW 160
GS+ +A HE K++ + K++ W+ L + AN SG+
Sbjct: 463 GSFGKALTNHEKKFKSTNDMEKLETWKMALNKVANLSGY 579
>TC228427 UP|Q8H6S7 (Q8H6S7) Resistance protein KR3, complete
Length = 2358
Score = 129 bits (325), Expect = 5e-31
Identities = 66/142 (46%), Positives = 94/142 (65%), Gaps = 1/142 (0%)
Frame = +1
Query: 19 KHEVFISFRSEDTRNNFTSHLNGALKRLDIRTYID-NNLNSGDEISTTLVRAIEEAELSV 77
+++VFI+FR EDTR FT HL+ AL IR ++D N++ GDEI TL AI+ + +++
Sbjct: 190 RYDVFINFRGEDTRFAFTGHLHKALCNKGIRAFMDENDIKRGDEIRATLEEAIKGSRIAI 369
Query: 78 IVFSKNYAASKFCLDELMKILECKRMKGKMVVPIFYDVDPTDVRNQRGSYAEAFAKHEKN 137
VFSK+YA+S FCLDEL IL C R K +V+P+FY VDP+DVR +GSYAE A+ E+
Sbjct: 370 TVFSKDYASSSFCLDELATILGCYREKTLLVIPVFYKVDPSDVRRLQGSYAEGLARLEER 549
Query: 138 SEEKIKVQEWRNGLMEAANYSG 159
++ W+ L + A +G
Sbjct: 550 FHP--NMENWKKALQKVAELAG 609
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,761,908
Number of Sequences: 63676
Number of extensions: 72735
Number of successful extensions: 426
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 373
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 377
length of query: 166
length of database: 12,639,632
effective HSP length: 90
effective length of query: 76
effective length of database: 6,908,792
effective search space: 525068192
effective search space used: 525068192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC134049.18


