

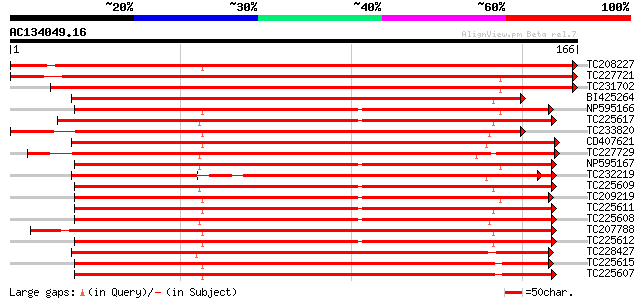
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.16 - phase: 0
(166 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208227 weakly similar to UP|Q9ZS32 (Q9ZS32) NL25, partial (17%) 251 1e-67
TC227721 similar to UP|Q8H0N7 (Q8H0N7) TIR-NBS disease resistanc... 226 5e-60
TC231702 weakly similar to UP|Q947E1 (Q947E1) Resistance gene an... 193 3e-50
BI425264 weakly similar to GP|28558777|g MRGH5 {Cucumis melo}, p... 139 5e-34
NP595166 R 14 protein [Glycine max] 139 8e-34
TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete 138 1e-33
TC233820 similar to UP|Q84ZV3 (Q84ZV3) R 4 protein, partial (15%) 137 2e-33
CD407621 135 7e-33
TC227729 135 9e-33
NP595167 R 13 protein [Glycine max] 135 1e-32
TC232219 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain... 134 2e-32
TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete 134 2e-32
TC209219 UP|Q9FVK2 (Q9FVK2) Resistance protein MG13 (Fragment), ... 133 3e-32
TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete 132 1e-31
TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete 132 1e-31
TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate re... 129 6e-31
TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete 129 8e-31
TC228427 UP|Q8H6S7 (Q8H6S7) Resistance protein KR3, complete 129 8e-31
TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete 127 2e-30
TC225607 homologue to UP|Q84ZV1 (Q84ZV1) R 9 protein, complete 125 7e-30
>TC208227 weakly similar to UP|Q9ZS32 (Q9ZS32) NL25, partial (17%)
Length = 697
Score = 251 bits (641), Expect = 1e-67
Identities = 124/167 (74%), Positives = 144/167 (85%), Gaps = 1/167 (0%)
Frame = +1
Query: 1 MAWSYSSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSG 59
MAWS SSSS+S N P +KHEVFISFR EDTR FTSHLN AL+RLDI+TY+DN+ L+ G
Sbjct: 52 MAWSSSSSSTS--NTPPQKHEVFISFRSEDTRKTFTSHLNAALERLDIKTYLDNNNLDRG 225
Query: 60 DEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSD 119
+EIPTTLVRAIEEAKLSVIVFSKNYA SKWCL+EL+KILE + K I++PVFYD+DPSD
Sbjct: 226 EEIPTTLVRAIEEAKLSVIVFSKNYADSKWCLDELLKILEFGRAKTLIIMPVFYDIDPSD 405
Query: 120 VRNQRGSYAEAFAKHENNFEGKIKVQEWRNGLLEAANYAGWDCNVNR 166
VRNQRG+YAEAF KHE F+ K K+QEWR GL+EAANY+GWDC+VNR
Sbjct: 406 VRNQRGTYAEAFDKHERYFQEKKKLQEWRKGLVEAANYSGWDCDVNR 546
>TC227721 similar to UP|Q8H0N7 (Q8H0N7) TIR-NBS disease resistance protein,
partial (19%)
Length = 1015
Score = 226 bits (575), Expect = 5e-60
Identities = 110/167 (65%), Positives = 136/167 (80%), Gaps = 1/167 (0%)
Frame = +2
Query: 1 MAWSYSSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGD 60
MAWS SSSS+ P +KHEVF+SFRGEDTR FT HL+ +L RL + TYID +L G+
Sbjct: 56 MAWSTSSSST-----PHQKHEVFLSFRGEDTRYTFTGHLHASLTRLQVNTYIDYNLQRGE 220
Query: 61 EIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDV 120
EI ++L+RAIEEAKLSV+VFSKNY SKWCL+EL+KILE K M+GQIV+P+FYD+DPS V
Sbjct: 221 EISSSLLRAIEEAKLSVVVFSKNYGNSKWCLDELLKILECKNMRGQIVLPIFYDIDPSHV 400
Query: 121 RNQRGSYAEAFAKHENNFEGKI-KVQEWRNGLLEAANYAGWDCNVNR 166
RNQ G+YAEAFAKHE + +G++ KVQ+WR L EAAN +GWDC+VNR
Sbjct: 401 RNQTGTYAEAFAKHEKHLQGQMDKVQKWRVALREAANLSGWDCSVNR 541
>TC231702 weakly similar to UP|Q947E1 (Q947E1) Resistance gene analog NBS7
(Fragment), partial (40%)
Length = 935
Score = 193 bits (491), Expect = 3e-50
Identities = 93/155 (60%), Positives = 117/155 (75%), Gaps = 1/155 (0%)
Frame = +1
Query: 13 NNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEE 72
+ P + H+VF+SFRGEDTR FTSHL AL RL ++TYIDN+L GDEI +L+RAI++
Sbjct: 16 SKTPQQVHDVFLSFRGEDTRYTFTSHLYAALTRLQVKTYIDNELERGDEISPSLLRAIDD 195
Query: 73 AKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFA 132
AK++VIVFS+NYA S+WCL+EL+KI+E K+ GQI+VPVFY VDP+ VR+Q GSY AFA
Sbjct: 196 AKVAVIVFSENYASSRWCLDELVKIMECKRKNGQIIVPVFYHVDPTHVRHQTGSYGHAFA 375
Query: 133 KHENNFEGKI-KVQEWRNGLLEAANYAGWDCNVNR 166
HE F G + KVQ WR L E AN +GWDC R
Sbjct: 376 MHEQRFVGNMNKVQTWRLVLGEVANISGWDCLTTR 480
>BI425264 weakly similar to GP|28558777|g MRGH5 {Cucumis melo}, partial (9%)
Length = 421
Score = 139 bits (351), Expect = 5e-34
Identities = 63/134 (47%), Positives = 97/134 (72%), Gaps = 1/134 (0%)
Frame = +1
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLSVI 78
+++VF+ FRGEDTR FT +L AL++ +RT+ D+ SGD+I +++AI+E+++S++
Sbjct: 10 RYDVFLCFRGEDTRYTFTGNLYAALRQARLRTFFDDGFKSGDQIFDVVLQAIQESRISIV 189
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+N+A S WCLEEL+KILE ++ K Q+V+P+FY +DPSDVR Q G Y E+ A+H+ F
Sbjct: 190 VLSENFASSSWCLEELVKILECRETKKQLVIPIFYRMDPSDVRRQTGCYGESLAQHQYEF 369
Query: 139 EG-KIKVQEWRNGL 151
KV+ W+ L
Sbjct: 370 RSDSEKVRNWQEAL 411
>NP595166 R 14 protein [Glycine max]
Length = 1926
Score = 139 bits (349), Expect = 8e-34
Identities = 70/142 (49%), Positives = 102/142 (71%), Gaps = 2/142 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SFRGEDTR FT +L AL I T+ D + L+ GDEI L +AI+E+++++
Sbjct: 34 YDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSKAIQESRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+EL+ IL K +G +V+PVFY+VDPSD+R+Q+GSY EA KH+ F
Sbjct: 214 VLSQNYAFSSFCLDELVTILHCKS-EGLLVIPVFYNVDPSDLRHQKGSYGEAMIKHQKRF 390
Query: 139 EGKI-KVQEWRNGLLEAANYAG 159
E K+ K+Q+WR L + A+ +G
Sbjct: 391 ESKMEKLQKWRMALKQVADLSG 456
>TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete
Length = 2697
Score = 138 bits (348), Expect = 1e-33
Identities = 72/148 (48%), Positives = 106/148 (70%), Gaps = 2/148 (1%)
Frame = +1
Query: 15 APLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEA 73
+P ++VF+SFRG DTR+ FT +L AL I T+ID+ +L GDEI L +AI+E+
Sbjct: 19 SPASIYDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQES 198
Query: 74 KLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAK 133
++++ V S+NYA S +CL+EL+ +L K+ KG +V+PVFY+VDPSDVR Q+GSY EA AK
Sbjct: 199 RIAITVLSQNYASSSFCLDELVTVLLCKR-KGLLVIPVFYNVDPSDVRQQKGSYGEAMAK 375
Query: 134 HENNFEG-KIKVQEWRNGLLEAANYAGW 160
H+ F+ K K+Q+WR L + A+ +G+
Sbjct: 376 HQKRFKAKKEKLQKWRMALHQVADLSGY 459
>TC233820 similar to UP|Q84ZV3 (Q84ZV3) R 4 protein, partial (15%)
Length = 471
Score = 137 bits (345), Expect = 2e-33
Identities = 73/153 (47%), Positives = 103/153 (66%), Gaps = 2/153 (1%)
Frame = +1
Query: 1 MAWSYSSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSG 59
MAW SSSSS N ++VF+SFRGEDTRN FT HL L+ I T+ID++ L G
Sbjct: 28 MAWGSRSSSSSSN------YDVFLSFRGEDTRNAFTGHLYNTLQSKGIHTFIDDEKLQRG 189
Query: 60 DEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSD 119
++I L++AI+++++++ V S++YA S +CL+EL IL + K +V+PVFY VDPSD
Sbjct: 190 EQITPALMKAIQDSRVAITVLSEHYASSSFCLDELATILHCDQRKRLLVIPVFYKVDPSD 369
Query: 120 VRNQRGSYAEAFAKHENNFE-GKIKVQEWRNGL 151
VR+Q+GSY EA AK E F+ K+Q W+ L
Sbjct: 370 VRHQKGSYGEALAKLERRFQHDPEKLQNWKMAL 468
>CD407621
Length = 571
Score = 135 bits (341), Expect = 7e-33
Identities = 68/145 (46%), Positives = 92/145 (62%), Gaps = 2/145 (1%)
Frame = -3
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSV 77
+++VFISFRG DTRN F HL L R I + D++ L G+ I L++AI++++LS+
Sbjct: 569 RYDVFISFRGPDTRNTFVDHLYAHLLRKGIFVFKDDEKLQKGESISAQLLQAIQDSRLSI 390
Query: 78 IVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENN 137
IVFSK YA S WCL+E+ I + K+ V PVFYDVDPS VR+Q G+Y AF H +
Sbjct: 389 IVFSKQYASSTWCLDEMAAIADCKQQSNPTVFPVFYDVDPSHVRHQNGAYEVAFVSHRSR 210
Query: 138 F-EGKIKVQEWRNGLLEAANYAGWD 161
F E KV W + + A+ AGWD
Sbjct: 209 FREDPDKVDRWARAMTDLAHSAGWD 135
>TC227729
Length = 1391
Score = 135 bits (340), Expect = 9e-33
Identities = 71/159 (44%), Positives = 106/159 (66%), Gaps = 3/159 (1%)
Frame = +2
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPT 64
+SSSSSF +++VF+SFRGEDTRN+FT L ALK+ I + D+ D+ G+ I
Sbjct: 149 TSSSSSF------EYDVFVSFRGEDTRNSFTGFLFEALKKQGIEAFKDDKDIRKGESIAP 310
Query: 65 TLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQR 124
L+RAIE + + ++VFSK+YA S WCL EL I + ++++P+FYDVDPS VR Q
Sbjct: 311 ELIRAIEGSHVFLVVFSKDYASSTWCLRELAHIWNCIQTSSRLLLPIFYDVDPSQVRKQS 490
Query: 125 GSYAEAFAKHE--NNFEGKIKVQEWRNGLLEAANYAGWD 161
G Y +AF++H+ + F+ K +++ WR L + A+ +GWD
Sbjct: 491 GDYEKAFSQHQQSSRFQEK-EIKTWREVLKQVASLSGWD 604
>NP595167 R 13 protein [Glycine max]
Length = 1926
Score = 135 bits (339), Expect = 1e-32
Identities = 70/143 (48%), Positives = 100/143 (68%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SFRG DTRN FT +L AL I T+ID+ +L GD+I L AI E+++++
Sbjct: 34 YDVFLSFRGLDTRNGFTGNLYKALGDRGIYTFIDDQELPRGDKITPALSNAINESRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+EL+ IL K +G +V+PVFY VDPSDVR+Q+GSY E KH+ F
Sbjct: 214 VLSENYAFSSFCLDELVTILHCKS-EGLLVIPVFYKVDPSDVRHQKGSYGETMTKHQKRF 390
Query: 139 EGKI-KVQEWRNGLLEAANYAGW 160
E K+ K++EWR L + A+ +G+
Sbjct: 391 ESKMEKLREWRMALQQVADLSGY 459
>TC232219 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (21%)
Length = 845
Score = 134 bits (338), Expect = 2e-32
Identities = 71/143 (49%), Positives = 96/143 (66%), Gaps = 1/143 (0%)
Frame = +2
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLSVI 78
+++VF+SF G DTR +FT L AL R +TY+++D GD+I + I +++LS+I
Sbjct: 401 RNDVFLSFCGRDTRYSFTGFLYNALSRSGFKTYMNDD---GDQISQS---TIGKSRLSII 562
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
VFSKNYA S CL+EL+ ILE KMK Q+V P+FY V+P D+R QR SY EA +HEN
Sbjct: 563 VFSKNYAHSSSCLDELLAILECMKMKNQLVWPIFYKVEPRDIRRQRNSYGEAMTEHENML 742
Query: 139 -EGKIKVQEWRNGLLEAANYAGW 160
+ KVQ+WR+ L EAA GW
Sbjct: 743 GKDSEKVQKWRSALFEAAXLXGW 811
Score = 83.6 bits (205), Expect = 4e-17
Identities = 39/102 (38%), Positives = 66/102 (64%), Gaps = 1/102 (0%)
Frame = +2
Query: 56 LNSGDEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDV 115
L GD+I T ++ A+E +++S++VFS +A S CL++L+ I K Q+++P+FYDV
Sbjct: 5 LRRGDKIATAILTAMEASRISIVVFSPYFASSTCCLDQLVHIHRCMNTKNQLILPIFYDV 184
Query: 116 DPSDVRNQRGSYAEAFAKHENNF-EGKIKVQEWRNGLLEAAN 156
D SDVR+Q ++ +A +H++ F + KV +W + L AN
Sbjct: 185 DQSDVRDQLNTFGQAMLQHQHRFGKSSDKVLQWSSVLSHVAN 310
>TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete
Length = 2688
Score = 134 bits (337), Expect = 2e-32
Identities = 71/143 (49%), Positives = 102/143 (70%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SFRG DTR+ FT +L AL I T ID+ +L GDEI L +AI+E+++++
Sbjct: 34 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+EL+ IL K +G +V+PVFY VDPSDVR+Q+GSY EA AKH+ F
Sbjct: 214 VLSQNYASSSFCLDELVTILHCKS-EGLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 390
Query: 139 EG-KIKVQEWRNGLLEAANYAGW 160
+ K K+Q+WR L + A+ +G+
Sbjct: 391 KAKKEKLQKWRMALKQVADLSGY 459
>TC209219 UP|Q9FVK2 (Q9FVK2) Resistance protein MG13 (Fragment), complete
Length = 1224
Score = 133 bits (335), Expect = 3e-32
Identities = 68/142 (47%), Positives = 101/142 (70%), Gaps = 2/142 (1%)
Frame = +3
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SFRG DTR FT +L AL T+ D D L+SG+EI L++AI+++++++I
Sbjct: 63 YDVFLSFRGTDTRYGFTGNLYKALCDKGFHTFFDEDKLHSGEEITPALLKAIQDSRVAII 242
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+EL+ I K+ +G +V+PVFY VDPS VR+Q+GSY EA KH+ F
Sbjct: 243 VLSENYAFSSFCLDELVTIFHCKR-EGLLVIPVFYKVDPSYVRHQKGSYGEAMTKHQERF 419
Query: 139 EGKI-KVQEWRNGLLEAANYAG 159
+ K+ K+QEWR L + A+ +G
Sbjct: 420 KDKMEKLQEWRMALKQVADLSG 485
>TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete
Length = 2709
Score = 132 bits (331), Expect = 1e-31
Identities = 68/143 (47%), Positives = 103/143 (71%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVI 78
++VF++FRGEDTR FT +L AL I T+ D D L+SGD+I L +AI+E+++++
Sbjct: 34 YDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSKAIQESRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+EL+ IL K+ +G +V+PVF++VDPS VR+ +GSY EA AKH+ F
Sbjct: 214 VLSQNYASSSFCLDELVTILHCKR-EGLLVIPVFHNVDPSAVRHLKGSYGEAMAKHQKRF 390
Query: 139 EG-KIKVQEWRNGLLEAANYAGW 160
+ K K+Q+WR L + A+ +G+
Sbjct: 391 KAKKEKLQKWRMALHQVADLSGY 459
>TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete
Length = 3084
Score = 132 bits (331), Expect = 1e-31
Identities = 69/143 (48%), Positives = 100/143 (69%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDN-DLNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SF G+DTR FT +L AL I T+ID+ +L GDEI L AI+E+++++
Sbjct: 34 YDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+EL+ IL K +G +V+PVFY VDPS VR+Q+GSY EA AKH+ F
Sbjct: 214 VLSQNYASSSFCLDELVTILHCKS-QGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRF 390
Query: 139 E-GKIKVQEWRNGLLEAANYAGW 160
+ K K+Q+WR L + A+ +G+
Sbjct: 391 KANKEKLQKWRMALHQVADLSGY 459
>TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate resistance
protein KR1, complete
Length = 3949
Score = 129 bits (324), Expect = 6e-31
Identities = 72/158 (45%), Positives = 103/158 (64%), Gaps = 4/158 (2%)
Frame = +1
Query: 7 SSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTT 65
SSSSSF+ ++VF+SFRGEDTR FT +L AL I T++D+ + GD+I +
Sbjct: 112 SSSSSFSYR--FSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQITSG 285
Query: 66 LVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRG 125
L +AIEE+++ +IV S+NYA S +CL EL IL+ K KG +++PVFY VDPSDVRN G
Sbjct: 286 LEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNHTG 465
Query: 126 SYAEAFAKHENNFEG---KIKVQEWRNGLLEAANYAGW 160
S+ +A HE F+ K++ W+ L + AN +G+
Sbjct: 466 SFGKALTNHEKKFKSTNDMEKLETWKMALNKVANLSGY 579
>TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete
Length = 2706
Score = 129 bits (323), Expect = 8e-31
Identities = 67/143 (46%), Positives = 99/143 (68%), Gaps = 2/143 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVI 78
++VF++FRG DTR FT +L AL I T+ D L+ G+EI L++AI+E+++++
Sbjct: 34 YDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLKAIQESRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V SKNYA S +CL+EL+ IL K +G +V+PVFY+VDPSDVR+Q+GSY AKH+ F
Sbjct: 214 VLSKNYASSSFCLDELVTILHCKS-EGLLVIPVFYNVDPSDVRHQKGSYGVEMAKHQKRF 390
Query: 139 EG-KIKVQEWRNGLLEAANYAGW 160
+ K K+Q+WR L + A+ G+
Sbjct: 391 KAKKEKLQKWRIALKQVADLCGY 459
>TC228427 UP|Q8H6S7 (Q8H6S7) Resistance protein KR3, complete
Length = 2358
Score = 129 bits (323), Expect = 8e-31
Identities = 68/142 (47%), Positives = 94/142 (65%), Gaps = 1/142 (0%)
Frame = +1
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYID-NDLNSGDEIPTTLVRAIEEAKLSV 77
+++VFI+FRGEDTR FT HL+ AL IR ++D ND+ GDEI TL AI+ +++++
Sbjct: 190 RYDVFINFRGEDTRFAFTGHLHKALCNKGIRAFMDENDIKRGDEIRATLEEAIKGSRIAI 369
Query: 78 IVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENN 137
VFSK+YA S +CL+EL IL + K +V+PVFY VDPSDVR +GSYAE A+ E
Sbjct: 370 TVFSKDYASSSFCLDELATILGCYREKTLLVIPVFYKVDPSDVRRLQGSYAEGLARLEER 549
Query: 138 FEGKIKVQEWRNGLLEAANYAG 159
F ++ W+ L + A AG
Sbjct: 550 FHP--NMENWKKALQKVAELAG 609
>TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete
Length = 2724
Score = 127 bits (319), Expect = 2e-30
Identities = 64/141 (45%), Positives = 96/141 (67%), Gaps = 1/141 (0%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVI 78
++VF+SFRGEDTR FT +L AL I T+ D D L+SG+EI L++AI+++++++
Sbjct: 34 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 213
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+++A S +CL+EL IL + G +V+PVFY V P DVR+Q+G+Y EA AKH+ F
Sbjct: 214 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 393
Query: 139 EGKIKVQEWRNGLLEAANYAG 159
K+ Q+W L + AN +G
Sbjct: 394 PDKL--QKWERALRQVANLSG 450
>TC225607 homologue to UP|Q84ZV1 (Q84ZV1) R 9 protein, complete
Length = 1063
Score = 125 bits (315), Expect = 7e-30
Identities = 62/142 (43%), Positives = 98/142 (68%), Gaps = 1/142 (0%)
Frame = +3
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVI 78
++VF++FRGEDTR FTS+L AL IRT+ D + L+SG+EI L++AI+++++++
Sbjct: 60 YDVFLNFRGEDTRYGFTSNLYRALSDKGIRTFFDEEKLHSGEEITPALLKAIKDSRIAIT 239
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+++A S +CL+EL I+ + G +++PVFY V PSDVR+Q+G+Y EA AKH+ F
Sbjct: 240 VLSEDFASSSFCLDELTSIVHCAQYNGMMIIPVFYKVYPSDVRHQKGTYGEALAKHKIRF 419
Query: 139 EGKIKVQEWRNGLLEAANYAGW 160
K Q W L + A+ +G+
Sbjct: 420 PEKF--QNWEMALRQVADLSGF 479
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,975,154
Number of Sequences: 63676
Number of extensions: 77710
Number of successful extensions: 462
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 411
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 413
length of query: 166
length of database: 12,639,632
effective HSP length: 90
effective length of query: 76
effective length of database: 6,908,792
effective search space: 525068192
effective search space used: 525068192
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC134049.16


