

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133864.8 - phase: 0
(509 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
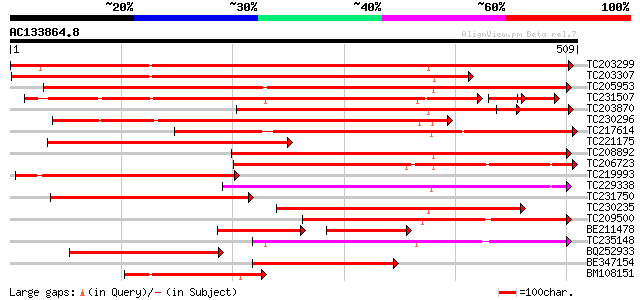
Score E
Sequences producing significant alignments: (bits) Value
TC203299 weakly similar to PDB|1CBG.0|1311386|1CBG Cyanogenic Be... 751 0.0
TC203307 weakly similar to UP|Q84L69 (Q84L69) P66 protein, parti... 578 e-165
TC205953 similar to UP|Q700B1 (Q700B1) Non-cyanogenic beta-gluco... 536 e-153
TC231507 UP|Q8S3J3 (Q8S3J3) Hydroxyisourate hydrolase, partial (... 353 e-104
TC203870 weakly similar to UP|Q7X9A9 (Q7X9A9) Beta-primeverosida... 335 4e-92
TC230296 similar to PIR|F86392|F86392 T1K7.7 protein - Arabidops... 332 3e-91
TC217614 similar to UP|Q7Y073 (Q7Y073) Latex cyanogenic beta glu... 317 1e-86
TC221175 weakly similar to UP|Q7X9A9 (Q7X9A9) Beta-primeverosida... 311 4e-85
TC208892 weakly similar to UP|Q40984 (Q40984) Amygdalin hydrolas... 307 6e-84
TC206723 similar to UP|Q8VWL8 (Q8VWL8) Beta-mannosidase enzyme, ... 293 2e-79
TC219993 similar to PDB|1CBG.0|1311386|1CBG Cyanogenic Beta-Gluc... 267 1e-71
TC229338 weakly similar to UP|O80690 (O80690) F8K4.3 protein, pa... 262 3e-70
TC231750 similar to UP|BGLS_TRIRP (P26204) Non-cyanogenic beta-g... 251 5e-67
TC230235 weakly similar to UP|Q945I3 (Q945I3) Prunasin hydrolase... 233 2e-61
TC209500 weakly similar to UP|Q43073 (Q43073) Prunasin hydrolase... 221 8e-58
BE211478 weakly similar to GP|15723332|gb prunasin hydrolase iso... 132 2e-56
TC235148 similar to UP|Q8S3J3 (Q8S3J3) Hydroxyisourate hydrolase... 212 4e-55
BQ252933 similar to SP|P26204|BGLS Non-cyanogenic beta-glucosida... 198 5e-51
BE347154 weakly similar to GP|15723332|gb prunasin hydrolase iso... 189 3e-48
BM108151 similar to GP|5030906|dbj| beta-glucosidase {Polygonum ... 181 5e-46
>TC203299 weakly similar to PDB|1CBG.0|1311386|1CBG Cyanogenic
Beta-Glucosidase Mol_id: 1; Molecule: Cyanogenic
Beta-Glucosidase; Chain: Null; Ec: 3.2.1.21. {Trifolium
repens;} , partial (80%)
Length = 1781
Score = 751 bits (1939), Expect = 0.0
Identities = 360/515 (69%), Positives = 423/515 (81%), Gaps = 9/515 (1%)
Frame = +1
Query: 1 MKAISHFLLYLF-SLATLLAVVTGTAS--QHVHPSHYAASFNRTLFPSDFLFGIGSSAYQ 57
+K S LL LF SLA LLA T ++ + PSH+ ++FNR+LFPS FLFGIGSSAYQ
Sbjct: 13 VKTQSASLLCLFLSLAILLASGTAASATPRSAVPSHHVSTFNRSLFPSTFLFGIGSSAYQ 192
Query: 58 IEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFS 117
EGA+++DGRGPSIWDT+T+QH EKI DHS+G++GADFYHRYK DIKI KEIGLDS+RFS
Sbjct: 193 AEGAASVDGRGPSIWDTYTRQHTEKIWDHSTGDMGADFYHRYKGDIKIAKEIGLDSFRFS 372
Query: 118 ISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLS 177
ISWSRIFP KGKGAVNP+GVKFYNNVI+E+LANGL PFVTLFHWD PQ+LEDEY GF S
Sbjct: 373 ISWSRIFP--KGKGAVNPLGVKFYNNVIDEILANGLKPFVTLFHWDFPQALEDEYGGFRS 546
Query: 178 PKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGD 237
PK+V DF YA+FCFKTFGDRVK+WVTLNEP+S+++NGY+GGT P RCSKYV NCS GD
Sbjct: 547 PKVVADFRGYANFCFKTFGDRVKYWVTLNEPLSFSLNGYNGGTFAPGRCSKYVANCSAGD 726
Query: 238 STTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRA 297
S+TEPYIV H+ +L+H +AA LYK KYQA QKG+IGIT THY+ P S S AD+KAASRA
Sbjct: 727 SSTEPYIVGHYLLLAHESAATLYKTKYQARQKGQIGITNPTHYFLPKSQSAADYKAASRA 906
Query: 298 LDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSI 357
LDF FGW++ P+ YG YP+SM SS+G+RLPKFTK E + +K S DFLGVNYYTTYYA+
Sbjct: 907 LDFFFGWYSDPVFYGDYPESMKSSVGSRLPKFTKAESEGLKNSIDFLGVNYYTTYYAEHA 1086
Query: 358 PPTYINMTYFTDMQANL------IHSNEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYI 411
P N T++TD+ A+L +H WL ++PKGIH L+ HIKD YKN P+YI
Sbjct: 1087EPVSANRTFYTDILASLSTERNGLHVGTPTDLNWLFIFPKGIHLLMAHIKDKYKNLPIYI 1266
Query: 412 TENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWD 471
TENG+ +SRNDSIPVN ARKD IRIRYHD HLKFLLQAIK+G N+KGYYAWSFSDS+EWD
Sbjct: 1267TENGMAESRNDSIPVNEARKDSIRIRYHDGHLKFLLQAIKEGVNLKGYYAWSFSDSFEWD 1446
Query: 472 AGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLK 506
AGYTVRFG+IYVD+ NNLKRYPK+SAFWLQKFLLK
Sbjct: 1447AGYTVRFGLIYVDYKNNLKRYPKFSAFWLQKFLLK 1551
>TC203307 weakly similar to UP|Q84L69 (Q84L69) P66 protein, partial (57%)
Length = 1268
Score = 578 bits (1491), Expect = e-165
Identities = 277/421 (65%), Positives = 325/421 (76%), Gaps = 6/421 (1%)
Frame = +3
Query: 2 KAISHFLLYLFSLATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGA 61
KAI L L L TLLA +A +V PSHYAA FNR++FPS FLFGIGS+AYQIEGA
Sbjct: 9 KAICFSFLCLIILVTLLAGSIESAPANVKPSHYAAPFNRSVFPSGFLFGIGSAAYQIEGA 188
Query: 62 SNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWS 121
+ IDGRGPSIWDT+TKQ P KI DHS G++ DFYHRYKSDIK++KE+GLDSYRFSISWS
Sbjct: 189 AAIDGRGPSIWDTYTKQQPGKIWDHSDGSLAIDFYHRYKSDIKMVKEVGLDSYRFSISWS 368
Query: 122 RIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIV 181
RIFP KGKGAVN +GVKFYN++INE++ANGL PFVTLFHWDLPQ+LEDEY GFL P+IV
Sbjct: 369 RIFP--KGKGAVNTLGVKFYNDLINEIIANGLKPFVTLFHWDLPQALEDEYGGFLKPEIV 542
Query: 182 KDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTE 241
+DF YADFCFKTFGDRVKHWVTLNEP Y++NGY GG P RCS YVG C GDS+TE
Sbjct: 543 EDFRNYADFCFKTFGDRVKHWVTLNEPYGYSVNGYSGGNFAPGRCSNYVGKCPAGDSSTE 722
Query: 242 PYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFL 301
PYIV HH IL+H AA YK KYQAHQKG+IG+T++T ++EP SNS AD KAA RALDF+
Sbjct: 723 PYIVNHHLILAHGAAVNCYKNKYQAHQKGQIGVTIVTFFFEPKSNSDADRKAARRALDFM 902
Query: 302 FGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTY 361
FGWFA+PIT+G YP+SM S +G+RLP TK + +KG YDFLG+N T + PPT
Sbjct: 903 FGWFANPITFGDYPESMRSLVGSRLPTCTKAHSEFLKGIYDFLGINVNTVNVVEYAPPTT 1082
Query: 362 INMTYFTDMQANLIHSNEE------WSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENG 415
N TYFTDM A L + + S WL +YP+GIH L+T+I D Y NPPV+ITENG
Sbjct: 1083TN*TYFTDMLAKLSSTRNDLPIVTPTSQSWLYIYPEGIHKLITYIMDNYYNPPVFITENG 1262
Query: 416 I 416
+
Sbjct: 1263V 1265
>TC205953 similar to UP|Q700B1 (Q700B1) Non-cyanogenic beta-glucosidase ,
partial (91%)
Length = 1776
Score = 536 bits (1381), Expect = e-153
Identities = 256/481 (53%), Positives = 335/481 (69%), Gaps = 7/481 (1%)
Frame = +3
Query: 31 PSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGN 90
P H AAS R FP+ F+FG GSSAYQ EGA+ GRGPSIWDTFT HPEKI D ++G+
Sbjct: 84 PVHDAASLTRNSFPAGFIFGAGSSAYQFEGAAKEGGRGPSIWDTFTHNHPEKIRDGANGD 263
Query: 91 IGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLA 150
+ D YHRYK D+KIMK++ LDSYRFSISW RI P K G VN G+ +YNN+INE+LA
Sbjct: 264 VAVDQYHRYKEDVKIMKDMNLDSYRFSISWPRILPKGKLSGGVNQEGINYYNNLINELLA 443
Query: 151 NGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVS 210
NG++P+VTLFHWDL Q+LEDEY G LS IV DF+ YAD CFK FGDRVK W TLNEP
Sbjct: 444 NGVLPYVTLFHWDLHQALEDEYSGLLSSHIVDDFQDYADLCFKEFGDRVKFWTTLNEPWL 623
Query: 211 YTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKG 270
++ GY G + P RC+ C GD+ TEPYIV H+ IL+HAAA +YK KYQAHQKG
Sbjct: 624 FSQGGYATGATAPGRCTG--PQCLGGDAGTEPYIVTHNQILAHAAAVHVYKTKYQAHQKG 797
Query: 271 KIGITLITHYYEPYS-NSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKF 329
KIGITL+++++ P + NS +D KAA RA+DF +GW+ P+T G YP++M + +G+RLPKF
Sbjct: 798 KIGITLVSNWFIPLAENSTSDIKAARRAIDFQYGWYMEPLTKGEYPKNMRALVGSRLPKF 977
Query: 330 TKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLIHSNE------EWSN 383
TK + K++ GS+DF+G+NYY++ Y +PP+ + TD + N ++
Sbjct: 978 TKWQAKLVNGSFDFIGLNYYSSGYINGVPPSNDKPNFLTDSRTNTSFERNGRPLGLRAAS 1157
Query: 384 YWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHL 443
W+ YP+G+ L+ + K+ Y NP +YITENG+ + + ++ V A D RI Y+ H
Sbjct: 1158VWIYFYPRGLLDLLLYTKEKYNNPLIYITENGMNEFNDPTLSVEEALMDIYRIDYYYRHF 1337
Query: 444 KFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKF 503
+L AIK GANVKG++AWSF D EW AG+TVRFG+ +VD+ + LKRYPK SA W + F
Sbjct: 1338FYLRSAIKAGANVKGFFAWSFLDCNEWFAGFTVRFGLNFVDYKDGLKRYPKLSAQWYKNF 1517
Query: 504 L 504
L
Sbjct: 1518L 1520
>TC231507 UP|Q8S3J3 (Q8S3J3) Hydroxyisourate hydrolase, partial (87%)
Length = 1461
Score = 353 bits (905), Expect(3) = e-104
Identities = 184/415 (44%), Positives = 257/415 (61%), Gaps = 4/415 (0%)
Frame = +1
Query: 14 LATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWD 73
LA L+ +V G A +++R FP DF+FG G+SAYQ+EGA+N DGR PSIWD
Sbjct: 58 LALLVNLVVGVLG--------ADNYSRDDFPLDFVFGSGTSAYQVEGAANKDGRTPSIWD 213
Query: 74 TFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAV 133
TF +G++ D YH+YK D+++M E GLD+YRFSISWSR+ P+ G+G V
Sbjct: 214 TFAYAG---YAHGENGDVACDGYHKYKEDVQLMLETGLDAYRFSISWSRLLPN--GRGPV 378
Query: 134 NPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFK 193
NP G+++ NN+INE+++NG+ P TL+++DLPQ LEDEY G++S I++DF YA+ F+
Sbjct: 379 NPKGLQYSNNLINELISNGIQPHATLYNFDLPQVLEDEYGGWISRDIIRDFTYYAEVEFR 558
Query: 194 TFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSK--YVGNCSTGDSTTEPYIVAHHFIL 251
FGDRV +W T+NEP + + GY G SPP RCS N + G+ST EPY+ HH +L
Sbjct: 559 EFGDRVLYWTTVNEPNVFALGGYDQGNSPPRRCSPPFCATNDTMGNSTYEPYLAVHHILL 738
Query: 252 SHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITY 311
SH++AA+LY KY+ Q G +GI++ T P +N+ D A+ RA DF GW P+ Y
Sbjct: 739 SHSSAARLYWRKYRDKQHGFVGISIYTFGIFPQTNTEKDRVASQRARDFFVGWIMEPLQY 918
Query: 312 GHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINM--TYFTD 369
G YP SM ++ G R+P FT E K +KGS+DF+GV +YT N + D
Sbjct: 919 GDYPISMKTNAGERIPAFTNHESKQVKGSFDFIGVIHYTNLNVSDNSDALKNQLRDFTAD 1098
Query: 370 MQANLIHSNEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSI 424
M AN I + +SN L+ P G+ + K Y NPP++I ENG + N S+
Sbjct: 1099MAAN-IFGEDLFSNEEYLITPWGLRQELNKFKLLYGNPPIFIHENGQRTASNSSL 1260
Score = 36.2 bits (82), Expect(3) = e-104
Identities = 18/38 (47%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Frame = +3
Query: 457 KGYYAWSFSDSYEWDAGYTVRFGIIYVDFVN-NLKRYP 493
+G +AW F D +E GY FG+ YVD + LKRYP
Sbjct: 1341 RGIFAWPFLDLFELLDGYKSSFGLYYVDRDDPQLKRYP 1454
Score = 31.2 bits (69), Expect(3) = e-104
Identities = 11/34 (32%), Positives = 22/34 (64%)
Frame = +2
Query: 431 KDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSF 464
K R++Y ++ +L A++D +N+KGY+ +F
Sbjct: 1262 KTWTRVKYLHGYIGSVLDALRDASNIKGYFRMAF 1363
>TC203870 weakly similar to UP|Q7X9A9 (Q7X9A9) Beta-primeverosidase ,
partial (41%)
Length = 1297
Score = 335 bits (858), Expect = 4e-92
Identities = 156/262 (59%), Positives = 193/262 (73%), Gaps = 6/262 (2%)
Frame = +3
Query: 204 TLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAK 263
T +P Y++NGY GG P RCS YVG C GDS+TEPYIV HH IL+H AA YK K
Sbjct: 57 TKQQPYGYSVNGYSGGNFAPGRCSNYVGKCPAGDSSTEPYIVNHHLILAHGAAVNCYKNK 236
Query: 264 YQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLG 323
YQAHQKG+IG+T++T ++EP SNS AD KAA RALDF+FGWFA+PIT+G YP+SM S +G
Sbjct: 237 YQAHQKGQIGVTIVTFFFEPKSNSDADRKAARRALDFMFGWFANPITFGDYPESMRSLVG 416
Query: 324 NRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANL------IHS 377
+RLP FTK + + +KGSYDFLG+NYYT+ + + PPT N TYFTDM A L +
Sbjct: 417 SRLPTFTKAQSESLKGSYDFLGINYYTSNFVEYAPPTTTNKTYFTDMLAKLSSTRNGVPI 596
Query: 378 NEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIR 437
WL +YP+GI+ L+T+I+D Y NPPVYITENG+ +S+NDS+ +N ARKDGIRIR
Sbjct: 597 GTPTPLSWLFIYPEGIYKLMTYIRDNYNNPPVYITENGVAESKNDSLAINEARKDGIRIR 776
Query: 438 YHDSHLKFLLQAIKDGANVKGY 459
YHD HLK LL AIKD +VK Y
Sbjct: 777 YHDGHLKSLLHAIKDRVSVKVY 842
Score = 107 bits (266), Expect = 2e-23
Identities = 49/69 (71%), Positives = 53/69 (76%)
Frame = +1
Query: 438 YHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSA 497
Y D L + KD NVKGYY WSFSDS+EWDAGYT RFGIIYVD+ NNL RYPK SA
Sbjct: 877 YADDFLDIPSLSCKDRVNVKGYYIWSFSDSFEWDAGYTARFGIIYVDYKNNLSRYPKSSA 1056
Query: 498 FWLQKFLLK 506
FWL+KFLLK
Sbjct: 1057FWLKKFLLK 1083
Score = 40.8 bits (94), Expect = 0.001
Identities = 18/29 (62%), Positives = 21/29 (72%), Gaps = 2/29 (6%)
Frame = +3
Query: 64 IDGRGPSIWDTFTKQHP--EKIGDHSSGN 90
IDGRGPSIWDT+TKQ P + +S GN
Sbjct: 21 IDGRGPSIWDTYTKQQPYGYSVNGYSGGN 107
>TC230296 similar to PIR|F86392|F86392 T1K7.7 protein - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (71%)
Length = 1103
Score = 332 bits (850), Expect = 3e-91
Identities = 169/369 (45%), Positives = 233/369 (62%), Gaps = 10/369 (2%)
Frame = +2
Query: 39 NRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHR 98
NR FP+ F FG SSA+Q EGA D RGPS+ DTF+ KI D S+ + D YHR
Sbjct: 2 NRENFPNGFAFGTASSAFQYEGAVKEDRRGPSVSDTFSHTFG-KIIDFSNADAAVDQYHR 178
Query: 99 YKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVT 158
Y+ DI++MK++ +D+YRFSISWSRIFP++ G+ +N GV YN +IN LA + P+VT
Sbjct: 179 YEQDIQLMKDMAMDAYRFSISWSRIFPNEYGQ--INQAGVDHYNKLINASLAKRIEPYVT 352
Query: 159 LFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHG 218
L+H DLPQ+LE++Y G+L+ I+ DF YA+ CF+ FGDRVKHW+T NEP ++ GY
Sbjct: 353 LYH*DLPQALENKYSGWLNASIIMDFATYAETCFQKFGDRVKHWITFNEPHTFATQGYDV 532
Query: 219 GTSPPARCSKYVGN-CSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLI 277
G P RCS + C G+S TEPYIVAH+ +LSHA A +Y+ KY+ Q G +G+
Sbjct: 533 GLQAPGRCSILLHLFCRAGNSATEPYIVAHNVLLSHATVADIYRKKYKKIQGGSLGVAFD 712
Query: 278 THYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKII 337
+YEP +N+ D AA RA DF GWF P+ +G YP SM + +G+RLPKF++ E ++
Sbjct: 713 VIWYEPLTNTKEDIDAAQRAQDFQLGWFLDPLMFGDYPSSMRTRVGSRLPKFSQSEAALV 892
Query: 338 KGSYDFLGVNYYTTYYAQSIPPTYINMTY---FTDMQANLIHSN------EEWSNYWLLV 388
KGS DF+G+N+YTT+YA+ I D A + N E S+ WL +
Sbjct: 893 KGSLDFVGINHYTTFYAKDNSTNLIGTLLHDSIADSGAVTLPFNGTKAISERASSIWLYI 1072
Query: 389 YPKGIHHLV 397
P+ + L+
Sbjct: 1073VPQSMKSLM 1099
>TC217614 similar to UP|Q7Y073 (Q7Y073) Latex cyanogenic beta glucosidase,
partial (73%)
Length = 1382
Score = 317 bits (811), Expect = 1e-86
Identities = 148/368 (40%), Positives = 227/368 (61%), Gaps = 7/368 (1%)
Frame = +3
Query: 149 LANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEP 208
L G+ P+VTL+HWDLP L + G+L+ +I++ F YAD CF +FGDRVK+W+T+NEP
Sbjct: 3 LERGIQPYVTLYHWDLPLHLHESMGGWLNKQIIEYFAVYADTCFASFGDRVKNWITINEP 182
Query: 209 VSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQ 268
+ +NGY P R +S EPY+ AHH IL+HAAA +Y++KY+ Q
Sbjct: 183 LQTAVNGYDVAIFAPGRRE---------NSLIEPYLAAHHQILAHAAAVSIYRSKYKDKQ 335
Query: 269 KGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPK 328
G++G + + E S+ + D AA+R LDF GWF HP+ YG YP+ M LG++LPK
Sbjct: 336 GGQVGFVVDCEWAEANSDKIEDKSAAARRLDFQLGWFLHPLYYGDYPEVMRERLGDQLPK 515
Query: 329 FTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLIHS-------NEEW 381
F++E+ KI+ + DF+G+N+YT+ + + Y+ + I E+
Sbjct: 516 FSEEDKKILLNALDFIGLNHYTSRFISHVTECAEENHYYKVQEMERIVEWEGGQAIGEKA 695
Query: 382 SNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDS 441
++ WL V P G+ ++ ++ Y P+++TENG+ ND++P++ D +R+RY
Sbjct: 696 ASEWLYVVPWGLRKILNYVSQKYAT-PIFVTENGMDDEDNDNLPLHEMLDDKLRVRYFKG 872
Query: 442 HLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQ 501
+L + QAIKDGA+V+GY+AWS D++EW GYT RFG++YVD+ N L R+PK SA+W
Sbjct: 873 YLASVAQAIKDGADVRGYFAWSLLDNFEWAQGYTKRFGLVYVDYKNGLSRHPKSSAYWFS 1052
Query: 502 KFLLKGKH 509
+FL G++
Sbjct: 1053RFLKAGEN 1076
>TC221175 weakly similar to UP|Q7X9A9 (Q7X9A9) Beta-primeverosidase ,
partial (43%)
Length = 681
Score = 311 bits (797), Expect = 4e-85
Identities = 139/220 (63%), Positives = 170/220 (77%)
Frame = +2
Query: 35 AASFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGAD 94
AAS NR+ FP+DF FG SSAYQ EGA+ G+GPSIWDTFT HP++I DHS+G++ D
Sbjct: 20 AASLNRSSFPADFFFGTASSAYQYEGAAREGGKGPSIWDTFTHSHPDRISDHSNGDVAID 199
Query: 95 FYHRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLI 154
YHRYK D+ +MK+IG ++YRFSISW RI P +G VN G+ +YNN+INE++ANG
Sbjct: 200 SYHRYKEDVAMMKDIGFNAYRFSISWPRILPRGNLQGGVNREGITYYNNLINELIANGQQ 379
Query: 155 PFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTIN 214
PF+TLFH D PQ+LEDEY GFLSPKI +DF YA+ CF+ FGDRVKHW+TLNEPV Y+
Sbjct: 380 PFITLFHSDFPQALEDEYGGFLSPKIEQDFANYAEVCFREFGDRVKHWITLNEPVLYSNG 559
Query: 215 GYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHA 254
GY G SPP RCSK+ NC+ GDSTTEPY+V HH IL+HA
Sbjct: 560 GYASGGSPPNRCSKWFANCTAGDSTTEPYVVTHHLILAHA 679
>TC208892 weakly similar to UP|Q40984 (Q40984) Amygdalin hydrolase isoform AH
I precursor , partial (49%)
Length = 1118
Score = 307 bits (787), Expect = 6e-84
Identities = 148/313 (47%), Positives = 206/313 (65%), Gaps = 8/313 (2%)
Frame = +1
Query: 200 KHWVTLNEPVSYTINGYHGGTSPPARCSKYVG-NCSTGDSTTEPYIVAHHFILSHAAAAK 258
KHW+TLNEP SY+ +GY G P RCS ++ NC+ GDS TEPY+V+HH +L+HAA+
Sbjct: 1 KHWITLNEPWSYSQHGYATGEMAPGRCSAWMNPNCNGGDSATEPYLVSHHQLLAHAASVH 180
Query: 259 LYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSM 318
+YK KYQ G IGITL ++Y P+S++ DHKA RA+DF +GWF P+T G YP+SM
Sbjct: 181 VYKTKYQTFFIGLIGITLNVNWYVPFSDNKLDHKATERAIDFQYGWFMDPLTTGDYPKSM 360
Query: 319 ISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPP-TYINMTYFTDMQANLIHS 377
+ RLPKFTKE+ K++ S+DF+G+NYY+ YA P + ++Y TD +N
Sbjct: 361 RFLVRARLPKFTKEQSKLLIDSFDFIGINYYSASYASDAPQLSNAKISYLTDSLSNSSFV 540
Query: 378 NE------EWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARK 431
+ ++ WL VYP+G ++ + K Y NP +YITENGI + + S+ + +
Sbjct: 541 RDGKPIGLNVASNWLYVYPRGFRDVLLYTKKKYNNPLIYITENGINEYDDSSLSLEESLL 720
Query: 432 DGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKR 491
D RI YH HL +L +AIK+G NVKGY+AWS D++EW GYTVRFG+ ++D+ N+LKR
Sbjct: 721 DIYRIDYHYRHLFYLQEAIKNGVNVKGYFAWSLLDNFEWHLGYTVRFGMNFIDYKNDLKR 900
Query: 492 YPKYSAFWLQKFL 504
Y K SA W + FL
Sbjct: 901 YSKLSALWFKDFL 939
>TC206723 similar to UP|Q8VWL8 (Q8VWL8) Beta-mannosidase enzyme, partial
(59%)
Length = 1132
Score = 293 bits (749), Expect = 2e-79
Identities = 151/318 (47%), Positives = 200/318 (62%), Gaps = 10/318 (3%)
Frame = +3
Query: 202 WVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYK 261
W+T NEP GY G P RCSK GNC+ G+S TEPYIVAH+ ILSHAAA + Y+
Sbjct: 3 WMTFNEPRVVAALGYDNGFFAPGRCSKEYGNCTAGNSGTEPYIVAHNLILSHAAAVQRYR 182
Query: 262 AKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISS 321
KYQ QKG+IGI L +YEP + S AD+ AA RA DF GWF HP+ YG YP+++ +
Sbjct: 183 EKYQEKQKGRIGILLDFVWYEPLTRSKADNFAAQRARDFHIGWFIHPLVYGEYPKTIQNI 362
Query: 322 LGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYA----QSIPPTYINMTYFTDMQANLIHS 377
+GNRLPKFT EE KI+KGS DF+G+N YTT++ QS P Y D A ++
Sbjct: 363 VGNRLPKFTSEEVKIVKGSIDFVGINQYTTFFIYDPHQSKPKV---PGYQMDWNAGFAYA 533
Query: 378 NE------EWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARK 431
++YWL P G++ + +IK+ Y NP V ++ENG+ N ++P +
Sbjct: 534 KNGVPIGPRANSYWLYNVPWGMYKSLMYIKERYGNPTVILSENGMDDPGNVTLPKGL--H 707
Query: 432 DGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKR 491
D RI Y+ +L L +A+ DGANV GY+AWS D++EW GYT RFGI+YVDF LKR
Sbjct: 708 DTTRINYYKGYLTQLKKAVDDGANVVGYFAWSLLDNFEWRLGYTSRFGIVYVDF-KTLKR 884
Query: 492 YPKYSAFWLQKFLLKGKH 509
YPK SA+W ++ + K K+
Sbjct: 885 YPKMSAYWFKQLITKKKY 938
>TC219993 similar to PDB|1CBG.0|1311386|1CBG Cyanogenic Beta-Glucosidase
Mol_id: 1; Molecule: Cyanogenic Beta-Glucosidase; Chain:
Null; Ec: 3.2.1.21. {Trifolium repens;} , partial (34%)
Length = 623
Score = 267 bits (682), Expect = 1e-71
Identities = 126/201 (62%), Positives = 149/201 (73%)
Frame = +3
Query: 6 HFLLYLFSLATLLAVVTGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNID 65
H L+ L L T L +T + P+ AS NR FP+ F+FG SSAYQ EG +N
Sbjct: 30 HVLVLLIVLVTSLITITQGV---ITPNPETASLNRNSFPTGFIFGTASSAYQYEGGANEG 200
Query: 66 GRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFP 125
GRGPSIWDTFT ++PEKI D SG++ D YHRYK D+ IMK++ LD+YRFSISWSRI P
Sbjct: 201 GRGPSIWDTFTHKYPEKIKDRDSGDVAVDSYHRYKEDVGIMKDMNLDAYRFSISWSRILP 380
Query: 126 SKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDFE 185
K G +N G+ +YNN+INE+LANGL PFVTLFHWDLPQSLEDEY GFLSP+IVKDF+
Sbjct: 381 EGKLSGGINQEGIDYYNNLINELLANGLKPFVTLFHWDLPQSLEDEYGGFLSPRIVKDFQ 560
Query: 186 AYADFCFKTFGDRVKHWVTLN 206
YAD CFK FGDRVKHW+TLN
Sbjct: 561 DYADLCFKEFGDRVKHWITLN 623
>TC229338 weakly similar to UP|O80690 (O80690) F8K4.3 protein, partial (47%)
Length = 1197
Score = 262 bits (669), Expect = 3e-70
Identities = 135/316 (42%), Positives = 180/316 (56%), Gaps = 3/316 (0%)
Frame = +1
Query: 192 FKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFIL 251
FK+FGDRVK+WVT NEP Y G PP RCS GNCS GDS EP++ AH+ IL
Sbjct: 4 FKSFGDRVKYWVTFNEPNYLVPLAYRLGIFPPLRCSSKFGNCSEGDSEKEPFVAAHNMIL 183
Query: 252 SHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITY 311
SHAAA LY+ KYQ Q G+IGI L +EP SNS AD A RA F W PI +
Sbjct: 184 SHAAAVDLYRNKYQTEQGGEIGIVLHCDSFEPLSNSTADKLATERAQSFSINWILDPILF 363
Query: 312 GHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTDMQ 371
G YP+ M LG LPK + + ++ DF+G+N+Y +YY + +
Sbjct: 364 GKYPKEMEMILGTTLPKSSSNDKAKLRQGLDFIGINHYASYYVRDCISSVCESGPGVSTT 543
Query: 372 ANLIHS---NEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNV 428
L E WL VYP G+ ++ ++KD Y N P++ITENG G + +
Sbjct: 544 EGLYQRTTIGELTPFDWLSVYPLGMKSILMYLKDRYNNTPMFITENGYGNLYDPDLTEEE 723
Query: 429 ARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNN 488
D RI + HL L+ AI++GA+V+GY+AWS D++EW G++VRFG+ +VDF +
Sbjct: 724 YLNDFKRIEFMSGHLDNLMAAIREGADVRGYFAWSLLDNFEWLYGFSVRFGLHHVDF-ST 900
Query: 489 LKRYPKYSAFWLQKFL 504
LKR PK SA W + F+
Sbjct: 901 LKRTPKLSAIWYEHFI 948
>TC231750 similar to UP|BGLS_TRIRP (P26204) Non-cyanogenic beta-glucosidase
precursor , partial (37%)
Length = 693
Score = 251 bits (641), Expect = 5e-67
Identities = 113/183 (61%), Positives = 139/183 (75%)
Frame = +1
Query: 37 SFNRTLFPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFY 96
S +R FP F+FG GSS+YQ EGA+ GR PS+WDTFT +P KI D S+G++ D Y
Sbjct: 136 SLSRNSFPEGFIFGAGSSSYQFEGAAKEGGREPSVWDTFTHNYPGKIMDRSNGDVAIDSY 315
Query: 97 HRYKSDIKIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPF 156
H YK D+ +MK++ LDSYRFSISWSRI P K G +N G+ +YNN+INE++ANG+ P
Sbjct: 316 HHYKEDVGMMKDMNLDSYRFSISWSRILPKGKLSGGINQEGINYYNNLINELIANGIQPL 495
Query: 157 VTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGY 216
VTLFHWDLPQ+LEDEY GFLSP+IVKDF YA+ CF FGDRVK+WVTLNEP SY+ +GY
Sbjct: 496 VTLFHWDLPQALEDEYGGFLSPRIVKDFRNYAELCFNEFGDRVKYWVTLNEPWSYSQHGY 675
Query: 217 HGG 219
G
Sbjct: 676 ANG 684
>TC230235 weakly similar to UP|Q945I3 (Q945I3) Prunasin hydrolase isoform PH
A (Fragment) , partial (44%)
Length = 702
Score = 233 bits (593), Expect = 2e-61
Identities = 115/231 (49%), Positives = 152/231 (65%), Gaps = 7/231 (3%)
Frame = +1
Query: 240 TEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPYSNSVADHKAASRALD 299
TEPY+V+HH +L+HAA ++YK KYQA Q G IGITL++H++ P SN+ D AA RA+D
Sbjct: 4 TEPYLVSHHQLLAHAAVVQVYKRKYQASQNGVIGITLVSHWFVPISNNKLDQNAAERAID 183
Query: 300 FLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPP 359
F+ GWF P+T G+YPQSM S +G RLPKF+K++ K I GS+DF+G+NYYT+ YA P
Sbjct: 184 FMLGWFLEPLTTGNYPQSMRSLVGKRLPKFSKQQTKSILGSFDFIGLNYYTSNYAIHEPQ 363
Query: 360 -TYINMTYFTDMQANL------IHSNEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYIT 412
Y TD QA L I ++ WL VYPKGI L+ ++K Y NP +YIT
Sbjct: 364 LRNAKPNYLTDFQAKLTTQRNGIPIGSNAASSWLYVYPKGIQELLLYVKKKYNNPLIYIT 543
Query: 413 ENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWS 463
ENGI + + ++ + D RI Y+ HL +L AIKDGANVKGY+ WS
Sbjct: 544 ENGIDEFNDPTLSIEEVLIDTYRIDYYYRHLFYLKSAIKDGANVKGYFVWS 696
>TC209500 weakly similar to UP|Q43073 (Q43073) Prunasin hydrolase isoform PH
I precursor , partial (35%)
Length = 954
Score = 221 bits (562), Expect = 8e-58
Identities = 111/247 (44%), Positives = 157/247 (62%), Gaps = 6/247 (2%)
Frame = +2
Query: 264 YQAHQKGKIGITLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLG 323
+QA QKG+I +TL + + P S S D +AA R L F++ WF P+ G YP M++ +G
Sbjct: 2 FQASQKGQIKVTLNSAWVVPLSQSKEDREAAYRGLAFMYDWFMEPLYSGTYPAVMVNRVG 181
Query: 324 NRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTD------MQANLIHS 377
RLPKFT+ EY ++KGSYDF+G+NYYT+ YA S P T FTD N +
Sbjct: 182 GRLPKFTRREYLMVKGSYDFIGLNYYTSTYATSSPCPRQRPTAFTDACVRFTTVRNGLLI 361
Query: 378 NEEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIR 437
+ ++ WL VYP GI L+ + K+ + NP +YITENGI + + + +N D RI
Sbjct: 362 GPKAASDWLYVYPPGIQGLLEYTKEKFNNPIIYITENGIDEVNDGKMLLN----DRTRID 529
Query: 438 YHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSA 497
Y HL +L +AI++G VKGY+AWS D++EW+AGY++RFG++YVD+ N LKR+ K SA
Sbjct: 530 YISHHLLYLQRAIRNGVRVKGYFAWSLLDNFEWNAGYSLRFGLVYVDYKNGLKRHRKRSA 709
Query: 498 FWLQKFL 504
W + FL
Sbjct: 710 LWFKIFL 730
>BE211478 weakly similar to GP|15723332|gb prunasin hydrolase isoform PH B
precursor {Prunus serotina}, partial (24%)
Length = 481
Score = 132 bits (331), Expect(2) = 2e-56
Identities = 57/79 (72%), Positives = 61/79 (77%)
Frame = +2
Query: 187 YADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVGNCSTGDSTTEPYIVA 246
YADFCFKTFGDRVKHWVTLNEP Y++NGY GG P RCS YVG C GDS+TEPYIV
Sbjct: 20 YADFCFKTFGDRVKHWVTLNEPYGYSVNGYSGGNFAPGRCSNYVGKCPAGDSSTEPYIVN 199
Query: 247 HHFILSHAAAAKLYKAKYQ 265
HH IL+H AA YK KYQ
Sbjct: 200 HHLILAHGAAVNCYKNKYQ 256
Score = 105 bits (263), Expect(2) = 2e-56
Identities = 47/76 (61%), Positives = 61/76 (79%)
Frame = +3
Query: 285 SNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFL 344
+ S AD KAA RALDF+FGWFA+PIT+G YP+SM S +G+RLP FTK + + +KGSYDFL
Sbjct: 249 NTSDADRKAARRALDFMFGWFANPITFGDYPESMRSLVGSRLPTFTKAQSESLKGSYDFL 428
Query: 345 GVNYYTTYYAQSIPPT 360
G+NYYT+ + + PPT
Sbjct: 429 GINYYTSDFVEYAPPT 476
>TC235148 similar to UP|Q8S3J3 (Q8S3J3) Hydroxyisourate hydrolase, partial
(45%)
Length = 988
Score = 212 bits (539), Expect = 4e-55
Identities = 117/291 (40%), Positives = 166/291 (56%), Gaps = 5/291 (1%)
Frame = +2
Query: 219 GTSPPARCSK--YVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITL 276
GTSPP RCS N + G+ST EPY+ HH +LSH++A +LY+ KY+ Q G +GI++
Sbjct: 14 GTSPPQRCSPPFCTTNSTRGNSTYEPYLAVHHILLSHSSAVRLYRRKYRDQQHGYVGISV 193
Query: 277 ITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKI 336
T + P ++S D A+ RA DFL GW P+ +G YP SM + G R+P FT E +
Sbjct: 194 YTFGFIPLTDSEKDKAASQRARDFLVGWIIEPLVHGDYPISMKKNAGARIPTFTTRESEQ 373
Query: 337 IKGSYDFLGVNYYTTYYAQSIPPTYIN--MTYFTDMQANLIHSNEEWSNYWLLVYPKGIH 394
+KGS DF+GV YY P DM A+LI+ + +S V P +
Sbjct: 374 LKGSSDFIGVIYYNNVNVTDNPDALKTPLRDILADMAASLIYLQDLFSEEEYPVTPWSLR 553
Query: 395 HLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGA 454
+ + + Y NPP++I ENG N S+ +D R++Y ++ +L A++DG+
Sbjct: 554 EELNNFQLHYGNPPIFIHENGQRTMSNSSL------QDVSRVKYLQGNIGGVLDALRDGS 715
Query: 455 NVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVN-NLKRYPKYSAFWLQKFL 504
N+KGY+AWSF D +E AGY FG+ YVD + LKRYPK SA W + FL
Sbjct: 716 NIKGYFAWSFLDLFELLAGYKSSFGLYYVDRDDPELKRYPKLSAKWYKWFL 868
>BQ252933 similar to SP|P26204|BGLS Non-cyanogenic beta-glucosidase precursor
(EC 3.2.1.21). [Creeping white clover] {Trifolium
repens}, partial (27%)
Length = 419
Score = 198 bits (503), Expect = 5e-51
Identities = 87/139 (62%), Positives = 111/139 (79%)
Frame = +2
Query: 54 SAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDS 113
++YQ EG +N G+GPSIWDTFT ++P+KI D S+G++ D YH YK D+ IMK + LD+
Sbjct: 2 ASYQYEGGANEGGKGPSIWDTFTHKYPDKIVDRSNGDVANDQYHHYKEDVGIMKYMNLDA 181
Query: 114 YRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYK 173
YRFSISWSRI P K G +N GVK+YNN+INE++ANGL PFVTLFHWDLPQ+LEDEY
Sbjct: 182 YRFSISWSRILPKGKLNGGINQEGVKYYNNLINELIANGLQPFVTLFHWDLPQALEDEYG 361
Query: 174 GFLSPKIVKDFEAYADFCF 192
GFL+P+I+ DF+ YA+ CF
Sbjct: 362 GFLNPRIINDFQDYAELCF 418
>BE347154 weakly similar to GP|15723332|gb prunasin hydrolase isoform PH B
precursor {Prunus serotina}, partial (22%)
Length = 484
Score = 189 bits (480), Expect = 3e-48
Identities = 85/131 (64%), Positives = 103/131 (77%)
Frame = +2
Query: 219 GTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLIT 278
G P RCS YVG C GDS+TEPYIV HH IL+H AA YK KYQAHQKG+IG+T++T
Sbjct: 92 GNFAPGRCSNYVGKCPAGDSSTEPYIVNHHLILAHGAAVNCYKNKYQAHQKGQIGVTIVT 271
Query: 279 HYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIK 338
++EP SNS AD KAA RALDF+FGWFA+PIT+G YP+SM S +G+RLP FTK + + +K
Sbjct: 272 FFFEPKSNSDADRKAARRALDFMFGWFANPITFGDYPESMRSLVGSRLPTFTKAQSESLK 451
Query: 339 GSYDFLGVNYY 349
GSYD LG+NYY
Sbjct: 452 GSYDXLGINYY 484
>BM108151 similar to GP|5030906|dbj| beta-glucosidase {Polygonum tinctorium},
partial (17%)
Length = 405
Score = 181 bits (460), Expect = 5e-46
Identities = 88/131 (67%), Positives = 103/131 (78%), Gaps = 4/131 (3%)
Frame = +3
Query: 104 KIMKEIGLDSYRFSISWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWD 163
KI KEIGLDS+RFSISWSRIFP KGKGAVNP+GVKFYNNVI+E+LANGL P VTLFHWD
Sbjct: 9 KIAKEIGLDSFRFSISWSRIFP--KGKGAVNPLGVKFYNNVIDEILANGLKPXVTLFHWD 182
Query: 164 LPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKH-WVTL---NEPVSYTINGYHGG 219
PQ+LEDEY GF PK+V DF Y FCF TFGDR ++ W +L N + +++ GY+GG
Sbjct: 183 FPQALEDEYGGFRXPKVVADFRGYXXFCFXTFGDRSQNIWGSLLXXNPXIXFSLXGYNGG 362
Query: 220 TSPPARCSKYV 230
P+RCSKYV
Sbjct: 363 XFAPSRCSKYV 395
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,428,943
Number of Sequences: 63676
Number of extensions: 403088
Number of successful extensions: 2014
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 1945
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1962
length of query: 509
length of database: 12,639,632
effective HSP length: 101
effective length of query: 408
effective length of database: 6,208,356
effective search space: 2533009248
effective search space used: 2533009248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC133864.8


