

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.6 - phase: 0
(368 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
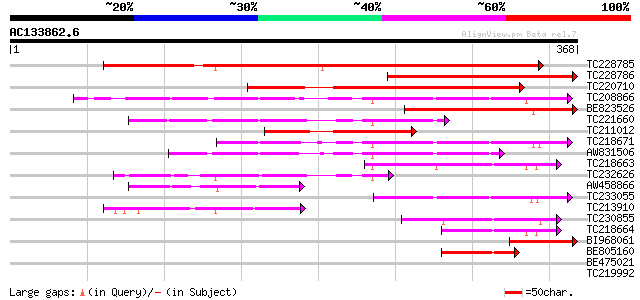
Score E
Sequences producing significant alignments: (bits) Value
TC228785 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g5... 454 e-128
TC228786 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g5... 230 7e-61
TC220710 similar to UP|Q8MLW3 (Q8MLW3) CG30386-PA, partial (7%) 224 4e-59
TC208866 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g1... 143 1e-34
BE823526 132 2e-31
TC221660 similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial (42%) 108 5e-24
TC211012 similar to GB|AAP42749.1|30984572|BT008736 At3g52060 {A... 107 1e-23
TC218671 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g1... 102 2e-22
AW831506 weakly similar to GP|4914330|gb| F14N23.16 {Arabidopsis... 91 1e-18
TC218663 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g3... 87 2e-17
TC232626 similar to GB|AAP37831.1|30725618|BT008472 At4g31350 {A... 77 2e-14
AW458866 76 3e-14
TC233055 weakly similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial... 73 2e-13
TC213910 weakly similar to UP|Q75M40 (Q75M40) Unknow protein, pa... 70 1e-12
TC230855 similar to GB|AAP37842.1|30725640|BT008483 At5g57270 {A... 58 8e-09
TC218664 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g3... 52 6e-07
BI968061 49 3e-06
BE805160 47 2e-05
BE475021 similar to GP|22535532|dbj P0431H09.21 {Oryza sativa (j... 40 0.002
TC219992 similar to UP|Q75M40 (Q75M40) Unknow protein, partial (... 40 0.002
>TC228785 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g52060
{Arabidopsis thaliana;} , partial (65%)
Length = 860
Score = 454 bits (1168), Expect = e-128
Identities = 218/290 (75%), Positives = 250/290 (86%), Gaps = 5/290 (1%)
Frame = +3
Query: 62 SHPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTL 121
S P FHLSSKNP+ KIAFLFLTN+DLHF+ LW+ FF TPS LFN+Y+H+DP VNLT
Sbjct: 6 SSPHSHFHLSSKNPSLKIAFLFLTNSDLHFSSLWDQFFSNTPSNLFNIYIHADPSVNLT- 182
Query: 122 LRSSNNYNPIF--KFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLH 179
+P+F KFISSK+T+R+SPTLISATRRLLA+A+LDD SNAYF +LSQ+CIPLH
Sbjct: 183 ----RPLSPLFINKFISSKRTFRSSPTLISATRRLLATALLDDPSNAYFALLSQHCIPLH 350
Query: 180 SFDYIYKSLFLSPTFDLTDSES---TQFGVRLKYKSFIEIINNGPRLWKRYTARGRYAMM 236
SF Y YKSLFLSPTFD D ES T+FG+RLKYKSF+EI+++ P+LWKRYT+RGRYAMM
Sbjct: 351 SFSYTYKSLFLSPTFDSQDPESSSSTRFGLRLKYKSFVEILSHAPKLWKRYTSRGRYAMM 530
Query: 237 PEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMED 296
PE+PFE FRVGSQFFTLTR+HALVVVKDRTLW+KFK+PCYRDDECYPEEHYFPTLLSM D
Sbjct: 531 PEIPFEAFRVGSQFFTLTRRHALVVVKDRTLWQKFKIPCYRDDECYPEEHYFPTLLSMAD 710
Query: 297 SDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESFLFA 346
DG T YTLT VNWTGTVNGHP+TY+P EVSPELILRLRKS +SES+LFA
Sbjct: 711 PDGCTKYTLTRVNWTGTVNGHPYTYRPTEVSPELILRLRKSNHSESYLFA 860
>TC228786 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g52060
{Arabidopsis thaliana;} , partial (36%)
Length = 826
Score = 230 bits (587), Expect = 7e-61
Identities = 106/123 (86%), Positives = 115/123 (93%)
Frame = +2
Query: 246 VGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTL 305
VGSQFFTLTR+HALVVVKDRTLWRKFK+PCYRDDECYPEEHYFPTLLSM D DG T YTL
Sbjct: 2 VGSQFFTLTRRHALVVVKDRTLWRKFKIPCYRDDECYPEEHYFPTLLSMADPDGCTKYTL 181
Query: 306 TNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESFLFARKFVPDCLEPLMGIAKSVI 365
T+VNWTGTVNGHP+TY+P E+SPELILRLRKS +SES+LFARKF PDCLEPLM IAKSVI
Sbjct: 182 TSVNWTGTVNGHPYTYRPTEISPELILRLRKSNHSESYLFARKFTPDCLEPLMRIAKSVI 361
Query: 366 FKD 368
F+D
Sbjct: 362 FRD 370
>TC220710 similar to UP|Q8MLW3 (Q8MLW3) CG30386-PA, partial (7%)
Length = 496
Score = 224 bits (572), Expect = 4e-59
Identities = 107/180 (59%), Positives = 130/180 (71%)
Frame = +1
Query: 155 LASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFI 214
LA+A+LDD N YF +LSQYCIPLHS + + LF +P +KSFI
Sbjct: 7 LAAALLDDPLNHYFALLSQYCIPLHSLQFTHNFLFKNP-----------------HKSFI 135
Query: 215 EIINNGPRLWKRYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVP 274
EI++N P L+ RYTARG +AM+PE+PF FRVGSQFF LTR+HA VVV+D LW KF++P
Sbjct: 136 EILSNEPNLFDRYTARGEHAMLPEIPFSSFRVGSQFFILTRRHARVVVRDILLWNKFRLP 315
Query: 275 CYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRL 334
C ++ CYPEEHYFPTLLSM+D +G TG+TLT VNWTG +GHPH Y EVSPELILRL
Sbjct: 316 CVTEEPCYPEEHYFPTLLSMQDPNGCTGFTLTRVNWTGCWDGHPHLYTAPEVSPELILRL 495
>TC208866 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;} , partial (78%)
Length = 1094
Score = 143 bits (361), Expect = 1e-34
Identities = 112/334 (33%), Positives = 163/334 (48%), Gaps = 10/334 (2%)
Frame = +2
Query: 42 LDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQT 101
++D LF A S P S+P K T KIAF+FLT L PLW FF+
Sbjct: 35 MNDTELFWRA---SFVPRIKSYPFK--------RTPKIAFMFLTKGPLPMAPLWEKFFRG 181
Query: 102 TPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILD 161
L+++YVHS P N SS Y + I S+ ++ A RRLLA+A+LD
Sbjct: 182 HEG-LYSIYVHSLPSYNADFSPSSVFYR---RQIPSQVAEWGMMSMCDAERRLLANALLD 349
Query: 162 DASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGP 221
SN +FI+LS+ CIPL +F +Y L+++ R +Y + GP
Sbjct: 350 -ISNEWFILLSESCIPLQNFSIVY--LYIA---------------RSRYSFMGAVDEPGP 475
Query: 222 RLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDD 279
RGRY M PE+ +R GSQ+F + R+ AL +V+D T + K K C +
Sbjct: 476 Y------GRGRYDGNMAPEINMSDWRKGSQWFEINRELALRIVEDNTYYPKLKEFC-KPH 634
Query: 280 ECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRL----- 334
+CY +EHYF T+L+ + +LT V+W+ HP T+ +++ E ++
Sbjct: 635 KCYVDEHYFQTMLTSNTPHLLANRSLTYVDWS-RGGAHPATFGKDDIKEEFFKKILQDQT 811
Query: 335 ---RKSTNSESFLFARKFVPDCLEPLMGIAKSVI 365
+S FLFARKF P+ L PL+ IA +
Sbjct: 812 CLYNNQPSSLCFLFARKFAPNALGPLLDIAPKAL 913
>BE823526
Length = 587
Score = 132 bits (333), Expect = 2e-31
Identities = 67/122 (54%), Positives = 77/122 (62%), Gaps = 10/122 (8%)
Frame = -1
Query: 257 HALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNG 316
HA +VV LW KF C R D CYPEE+YFPT LSM D G TLT+VNWTG V+G
Sbjct: 587 HARLVVSXXVLWSKFDAXCVRFDSCYPEENYFPTXLSMWDPQGCVPATLTHVNWTGRVDG 408
Query: 317 HPHTYQPEEVSPELILRLRKST----------NSESFLFARKFVPDCLEPLMGIAKSVIF 366
HP TY+ EV PELI R+R+ S+ FLFARKF PD L+PLM IA VIF
Sbjct: 407 HPRTYEAWEVGPELIRRMREDRPRYGDGNSDGRSDPFLFARKFAPDALQPLMRIANGVIF 228
Query: 367 KD 368
+D
Sbjct: 227 RD 222
>TC221660 similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial (42%)
Length = 545
Score = 108 bits (269), Expect = 5e-24
Identities = 76/210 (36%), Positives = 106/210 (50%), Gaps = 2/210 (0%)
Frame = +2
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISS 137
K+AF+FLT L PLW FF S LFN+Y+HS PR L + SS Y + I S
Sbjct: 5 KVAFMFLTRGPLPMLPLWERFFHGH-SSLFNIYIHSPPRFLLNVSHSSPFY---LRHIPS 172
Query: 138 KKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLT 197
+ + TL A RRLLA+A+LD SN F++LS+ CIP+++F +Y+ L S
Sbjct: 173 QDVSWGTVTLADAERRLLANALLD-FSNERFVLLSESCIPVYNFPTVYRYLTNSSL---- 337
Query: 198 DSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFTLTR 255
SF+E + R RGRY+ M+P + +R GSQ+F L R
Sbjct: 338 --------------SFVESYDEPTRY-----GRGRYSRNMLPHIQLRHWRKGSQWFELNR 460
Query: 256 KHALVVVKDRTLWRKFKVPCYRDDECYPEE 285
A+ +V D + F+ C CYP+E
Sbjct: 461 ALAVYIVSDTNYYSLFRKYC--KPACYPDE 544
>TC211012 similar to GB|AAP42749.1|30984572|BT008736 At3g52060 {Arabidopsis
thaliana;} , partial (20%)
Length = 326
Score = 107 bits (266), Expect = 1e-23
Identities = 53/99 (53%), Positives = 67/99 (67%)
Frame = +1
Query: 166 AYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRLWK 225
+YF +LSQ+CIPLHS + + LF +PT +KSFIEI++N P L+
Sbjct: 4 SYFALLSQHCIPLHSLQFTHNFLFKNPTHP--------------HKSFIEILSNEPNLFD 141
Query: 226 RYTARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKD 264
RYTARG +AM+PEVPF FRVGSQFF LTR+HA VV+D
Sbjct: 142 RYTARGEHAMLPEVPFSSFRVGSQFFILTRRHARTVVRD 258
Score = 39.3 bits (90), Expect = 0.003
Identities = 19/50 (38%), Positives = 25/50 (50%)
Frame = +2
Query: 237 PEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEH 286
P P R S TR V+ + LW KF++PC ++ CYPEEH
Sbjct: 185 PSPPSAWGRSSSSSLAATRARWCVI*R---LWNKFRLPCVTEEPCYPEEH 325
>TC218671 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;} , partial (46%)
Length = 899
Score = 102 bits (255), Expect = 2e-22
Identities = 80/240 (33%), Positives = 123/240 (50%), Gaps = 9/240 (3%)
Frame = +2
Query: 135 ISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTF 194
I S + + +I A RRLLA+A++D SN F++LS+ CIPL +F IY L
Sbjct: 8 IPSXEVEWGNVNMIEAERRLLANALVD-ISNQRFVLLSESCIPLFNFSTIYTYLM----- 169
Query: 195 DLTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRYA--MMPEVPFEKFRVGSQFFT 252
STQ +++ +++ + RGRY+ M+PE+ ++R GSQ+F
Sbjct: 170 -----NSTQ--------NYVMAVDDPSSV-----GRGRYSIQMLPEISLNQWRKGSQWFE 295
Query: 253 LTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTG 312
+ R AL VV DR + F+ C CY +EHY PT +S++ +G + +LT V+W+
Sbjct: 296 MDRDLALEVVSDRKYFPVFQDYC--KGSCYADEHYLPTYVSIKFWEGNSNRSLTWVDWS- 466
Query: 313 TVNGHPHTYQPEEVSPELILRLRKST---NSES----FLFARKFVPDCLEPLMGIAKSVI 365
HP + E++ + + LR N +S FLFARKF P + L IA V+
Sbjct: 467 KGGPHPTKFLRSEITVKFLESLRDQKCEYNGDSINVCFLFARKFAPGSVSKLTKIAPMVM 646
>AW831506 weakly similar to GP|4914330|gb| F14N23.16 {Arabidopsis thaliana},
partial (44%)
Length = 589
Score = 90.5 bits (223), Expect = 1e-18
Identities = 71/220 (32%), Positives = 103/220 (46%), Gaps = 2/220 (0%)
Frame = +2
Query: 104 SKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLASAILDDA 163
S LF++Y+H+ PR L + SS Y + I S+ + TL A RRLLA A+L
Sbjct: 14 SSLFSIYIHAPPRCTLNISHSSPFY---LRNIPSQDVSWGTFTLADAHRRLLADALLH-F 181
Query: 164 SNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFDLTDSESTQFGVRLKYKSFIEIINNGPRL 223
SN F++LS+ CIP + Y L T + Y S +E + R
Sbjct: 182 SNERFLLLSETCIPGYDLPTAYTDL-------------THY-----YLSCVESYDEPTRY 307
Query: 224 WKRYTARGRYA--MMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDEC 281
RGRY+ M+ + +R GSQ+ L R A+ +V D + F+ C C
Sbjct: 308 -----GRGRYSRHMLAHIHLRHWRKGSQWLELNRSLAVYIVSDTKYYSLFRKYC--KPAC 466
Query: 282 YPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTY 321
YP+EHY PT L M + T+T V+W+ + HP T+
Sbjct: 467 YPDEHYIPTFLHMFHGSLNSNRTVTWVDWS-MLGAHPATF 583
>TC218663 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g31350
{Arabidopsis thaliana;} , partial (47%)
Length = 915
Score = 86.7 bits (213), Expect = 2e-17
Identities = 50/156 (32%), Positives = 81/156 (51%), Gaps = 28/156 (17%)
Frame = +2
Query: 231 GRYA--MMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPC---YRDDECYPEE 285
GRY+ M+PEV + FR G+Q+F + R+HA++V+ D + KF+ C C +E
Sbjct: 44 GRYSDHMLPEVEVKDFRKGAQWFAMKRQHAIIVMADNLYYSKFRSYCQPGLEGKNCIADE 223
Query: 286 HYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRL----------- 334
HY PT M D G+ ++LT+V+W+ HP +Y+ ++V+ EL+ +
Sbjct: 224 HYLPTFFQMVDPGGIANWSLTHVDWSER-KWHPKSYRAQDVTYELLKNITSIDVSMHVTS 400
Query: 335 --RKSTNS----------ESFLFARKFVPDCLEPLM 358
+K S +LFARKF P+ L+ L+
Sbjct: 401 DEKKEVQSWPCLWNGIQKPCYLFARKFTPETLDSLL 508
>TC232626 similar to GB|AAP37831.1|30725618|BT008472 At4g31350 {Arabidopsis
thaliana;} , partial (44%)
Length = 888
Score = 76.6 bits (187), Expect = 2e-14
Identities = 62/187 (33%), Positives = 92/187 (49%), Gaps = 5/187 (2%)
Frame = +2
Query: 68 FHLSSKNPTFKIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNN 127
+++ +K P K+AFLFL+ L F LW++FFQ K F+VYVHS +
Sbjct: 110 YYMHTKKP--KVAFLFLSPGSLPFEKLWHMFFQGHEGK-FSVYVHSSKE-------KPTH 259
Query: 128 YNPIF--KFISSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIY 185
+ F + I S+ ++ A RRLLA A+LD N +F++LS+ CIP+ F+++Y
Sbjct: 260 VSSFFVGREIHSEPVGWGKISMAEAERRLLAHALLDP-DNQHFVLLSESCIPVRRFEFVY 436
Query: 186 KSLFLSPTFDLTDSESTQFGVRLKYKSFIE-IINNGPRLWKRYTARGRYA--MMPEVPFE 242
L L+ SFI+ ++ GP GRY M+PEV +
Sbjct: 437 NYLLLTNV------------------SFIDSYVDPGPH------GNGRYIEHMLPEVEKK 544
Query: 243 KFRVGSQ 249
FR GSQ
Sbjct: 545 DFRKGSQ 565
>AW458866
Length = 388
Score = 75.9 bits (185), Expect = 3e-14
Identities = 47/116 (40%), Positives = 67/116 (57%), Gaps = 2/116 (1%)
Frame = +2
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFK--FI 135
K+AF+FLT + PLW FF+ +++YVHS+P N S +P+FK I
Sbjct: 53 KVAFMFLTRGPVFLAPLWEQFFKGHEG-FYSIYVHSNPSYN-----GSRPESPVFKGRRI 214
Query: 136 SSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLS 191
SK+ + +I A RRLLA+A++ D SN F++LS+ CIPL +F IY L S
Sbjct: 215 PSKEVEWGNVNMIEAERRLLANALV-DISNQRFVLLSESCIPLFNFSTIYTYLMNS 379
>TC233055 weakly similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial (38%)
Length = 718
Score = 73.2 bits (178), Expect = 2e-13
Identities = 45/138 (32%), Positives = 69/138 (49%), Gaps = 9/138 (6%)
Frame = +2
Query: 237 PEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMED 296
P + +R G Q+F L R A+ +V D + F+ C CYP+EHY PT L M
Sbjct: 101 PHIXXRHWRKGXQWFELNRSLAVYIVSDTKYYSLFRKYC--KPACYPDEHYIPTFLHMFH 274
Query: 297 SDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKS-----TNSE----SFLFAR 347
+ T+T V+W+ + HP T+ ++ + +R + NSE +LFAR
Sbjct: 275 GSLNSNRTVTWVDWS-MLGPHPATFGRANITAAFLQSIRNNGSLCPYNSEMTSICYLFAR 451
Query: 348 KFVPDCLEPLMGIAKSVI 365
KF P LEPL+ ++ V+
Sbjct: 452 KFDPSALEPLLNLSSEVM 505
>TC213910 weakly similar to UP|Q75M40 (Q75M40) Unknow protein, partial (29%)
Length = 589
Score = 70.5 bits (171), Expect = 1e-12
Identities = 53/153 (34%), Positives = 75/153 (48%), Gaps = 22/153 (14%)
Frame = +1
Query: 62 SHPSKF--FHLSSK----NPTFKIAFL-------------FLTNTDLHFTPLWNLFFQTT 102
SH S F F LSS NP F+ FL FL +L LW+ FFQ
Sbjct: 91 SHASSFWLFFLSSDSTLPNPQFQFQFLASSSMALPRSLSLFLVRRNLPLDFLWDAFFQNG 270
Query: 103 PSKLFNVYVHSDPRVNLTLLRSSNNYNPIF---KFISSKKTYRASPTLISATRRLLASAI 159
F++YVHS P +L S + +F + +S + ++I A R LLA+A
Sbjct: 271 DVSRFSIYVHSAPGF---VLDESTTRSQLFYGRQISNSIQVLWGESSMIQAERLLLAAA- 438
Query: 160 LDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSP 192
L+D +N F++LS C+PL++F Y+Y L SP
Sbjct: 439 LEDHANQRFVLLSDSCVPLYNFSYVYNYLMASP 537
>TC230855 similar to GB|AAP37842.1|30725640|BT008483 At5g57270 {Arabidopsis
thaliana;} , partial (24%)
Length = 418
Score = 57.8 bits (138), Expect = 8e-09
Identities = 35/131 (26%), Positives = 61/131 (45%), Gaps = 27/131 (20%)
Frame = +3
Query: 255 RKHALVVVKDRTLWRKFKVPCYRDDE----CYPEEHYFPTLLSMEDSDGVTGYTLTNVNW 310
R+HA++V+ D + KFK C + E CY +EHY PT M D G+ +++T V+W
Sbjct: 6 RQHAIIVMADSLYFTKFKHHCRPNMEGNRNCYADEHYLPTFFIMLDPGGIANWSVTYVDW 185
Query: 311 TGTVNGHPHTYQPEEVSPELILRLRKSTNSESF-----------------------LFAR 347
+ HP +++ +++ +++ + S F LFAR
Sbjct: 186 SQR-KWHPRSFRARDITYQVMKNIAYIDESPHFTSDAKRTVVITPCMLNGSKRSCYLFAR 362
Query: 348 KFVPDCLEPLM 358
KF P+ + L+
Sbjct: 363 KFFPEAQDNLI 395
>TC218664 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g31350
{Arabidopsis thaliana;} , partial (29%)
Length = 673
Score = 51.6 bits (122), Expect = 6e-07
Identities = 29/101 (28%), Positives = 49/101 (47%), Gaps = 23/101 (22%)
Frame = +3
Query: 281 CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRL------ 334
C +EHY PT M D G+ ++LT+V+W+ HP +Y+ ++V+ EL+ +
Sbjct: 12 CIXDEHYLPTFFQMVDPGGIANWSLTHVDWSER-KWHPKSYRAQDVTYELLKNITSIDVS 188
Query: 335 -------RKSTNS----------ESFLFARKFVPDCLEPLM 358
+K S +LFARKF P+ ++ L+
Sbjct: 189 VHVTSDEKKEVQSWPCLWNGIQKPCYLFARKFTPETMDSLL 311
>BI968061
Length = 426
Score = 49.3 bits (116), Expect = 3e-06
Identities = 26/45 (57%), Positives = 34/45 (74%), Gaps = 1/45 (2%)
Frame = -3
Query: 325 EVSPELILRLRK-STNSESFLFARKFVPDCLEPLMGIAKSVIFKD 368
EV PELILR + +++S +LFARKF P+CL PLM IA VIF++
Sbjct: 415 EVWPELILRXXE*NSSSCVYLFARKFAPECLTPLMQIADDVIFRE 281
>BE805160
Length = 351
Score = 46.6 bits (109), Expect = 2e-05
Identities = 19/51 (37%), Positives = 32/51 (62%)
Frame = +2
Query: 281 CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELI 331
C +EHY PT M D G+ ++LT+V+W+ HP +Y+ ++V+ EL+
Sbjct: 8 CIADEHYLPTFFQMVDPGGIANWSLTHVDWSER-KWHPKSYRAQDVTYELL 157
>BE475021 similar to GP|22535532|dbj P0431H09.21 {Oryza sativa (japonica
cultivar-group)}, partial (17%)
Length = 178
Score = 40.0 bits (92), Expect = 0.002
Identities = 15/33 (45%), Positives = 24/33 (72%)
Frame = +1
Query: 160 LDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSP 192
L+D +N F++LS C+PL++F Y+Y L +SP
Sbjct: 1 LEDPANQRFVLLSDSCVPLYNFSYMYNYLMVSP 99
>TC219992 similar to UP|Q75M40 (Q75M40) Unknow protein, partial (33%)
Length = 937
Score = 39.7 bits (91), Expect = 0.002
Identities = 32/127 (25%), Positives = 55/127 (43%), Gaps = 28/127 (22%)
Frame = +3
Query: 268 WRKFKVPC--YRDDECYPEEHYFPTLLSMED-SDGVTGYTLTNVNWTGTVN-------GH 317
WR +P + C P+EHY TLL+ + + +T +LT+ +W + + H
Sbjct: 48 WRAHYIPADTSKVHNCIPDEHYVQTLLAQKGLEEEITRRSLTHTSWDISNSREHERRGWH 227
Query: 318 PHTYQPEEVSPELILRLRKSTN------------------SESFLFARKFVPDCLEPLMG 359
P TY+ + +P L+ +++ N S FLFARKF L+
Sbjct: 228 PVTYKYSDATPMLLKFVKEIDNIYYETEYRREWCSSKGKPSTCFLFARKFTRTAALRLLN 407
Query: 360 IAKSVIF 366
++ +F
Sbjct: 408 MSVLGVF 428
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,820,428
Number of Sequences: 63676
Number of extensions: 344037
Number of successful extensions: 3051
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 2938
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3017
length of query: 368
length of database: 12,639,632
effective HSP length: 99
effective length of query: 269
effective length of database: 6,335,708
effective search space: 1704305452
effective search space used: 1704305452
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133862.6


