

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.9 - phase: 0 /pseudo
(562 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
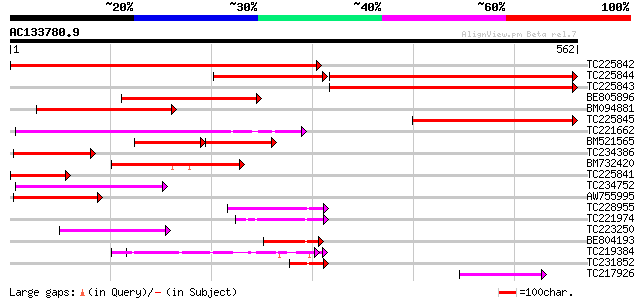
Score E
Sequences producing significant alignments: (bits) Value
TC225842 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respo... 539 e-153
TC225844 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respo... 273 e-119
TC225843 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respo... 285 3e-77
BE805896 similar to GP|9279671|dbj transport inhibitor response-... 233 2e-61
BM094881 similar to PIR|T48087|T48 transport inhibitor response ... 206 2e-53
TC225845 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respo... 181 1e-45
TC221662 weakly similar to UP|Q6TDU2 (Q6TDU2) Coronatine-insensi... 158 7e-39
BM521565 similar to PIR|T48087|T48 transport inhibitor response ... 105 2e-36
TC234386 similar to PIR|T48087|T48087 transport inhibitor respon... 129 3e-30
BM732420 weakly similar to GP|13249030|gb F-box containing prote... 127 2e-29
TC225841 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respo... 121 7e-28
TC234752 weakly similar to UP|Q6TDU2 (Q6TDU2) Coronatine-insensi... 117 2e-26
AW755995 similar to PIR|T48087|T480 transport inhibitor response... 112 6e-25
TC228955 weakly similar to PIR|T48087|T48087 transport inhibitor... 75 6e-14
TC221974 similar to UP|Q9AUH6 (Q9AUH6) F-box containing protein ... 69 7e-12
TC223250 similar to UP|Q6TDU2 (Q6TDU2) Coronatine-insensitive 1,... 56 4e-08
BE804193 similar to GP|13249030|gb| F-box containing protein TIR... 56 4e-08
TC219384 51 1e-06
TC231852 similar to PIR|T48087|T48087 transport inhibitor respon... 49 8e-06
TC217926 similar to PIR|T48087|T48087 transport inhibitor respon... 48 1e-05
>TC225842 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein (At3g26830), partial (54%)
Length = 1777
Score = 539 bits (1388), Expect = e-153
Identities = 263/309 (85%), Positives = 286/309 (92%)
Frame = +1
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEH+FDYVVSHSDRN+LSLVCKSWYRIER TRQRVFIGNCYSI+PERL++RF
Sbjct: 676 MNYFPDEVIEHIFDYVVSHSDRNALSLVCKSWYRIERCTRQRVFIGNCYSITPERLIQRF 855
Query: 61 PDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELL 120
P LKSLTLKGKPHFADFSLVP+ WGGFV+PW+EALAK++VGLEELRLKRMVVSDESLELL
Sbjct: 856 PGLKSLTLKGKPHFADFSLVPYDWGGFVHPWVEALAKSRVGLEELRLKRMVVSDESLELL 1035
Query: 121 SRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTS 180
SRSF +FKSLVLVSCEGF+TDGLAA+AANCR LRELDLQENEVEDHKGQWLSCFP++CTS
Sbjct: 1036SRSFTHFKSLVLVSCEGFSTDGLAALAANCRFLRELDLQENEVEDHKGQWLSCFPDNCTS 1215
Query: 181 LVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSF 240
LVSLNFACLKG+++LGALER V+RSPNLKSL+LNRSVPVDALQRI+ RAPQL DLGIGS
Sbjct: 1216LVSLNFACLKGEVSLGALERFVARSPNLKSLKLNRSVPVDALQRIMMRAPQLSDLGIGSL 1395
Query: 241 FHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGI 300
HD S+AY K TILKCKSITSLSGFLEVAP LAAIYPIC NLTSLNLSYAAGI G
Sbjct: 1396VHDPESEAYIKLKNTILKCKSITSLSGFLEVAPHCLAAIYPICPNLTSLNLSYAAGIQGS 1575
Query: 301 ELIKLIRHC 309
L+KLI HC
Sbjct: 1576ALVKLIHHC 1602
>TC225844 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein (At3g26830), partial (61%)
Length = 1347
Score = 273 bits (699), Expect(2) = e-119
Identities = 153/245 (62%), Positives = 170/245 (68%)
Frame = +1
Query: 318 D*VL*LAHVKNCKN*EFSHQLHLETKQLLPKKDL*RYQWDVLSFTHYSTSVTR*PMLLS* 377
D +L*L HVK CKN* H LE +Q KK +YQW LSFTH S S TR* ML S*
Sbjct: 388 DLML*LLHVKTCKN*GCFHLFPLEIQQPSQKKGWLQYQWVALSFTHCSISATR*QMLHS* 567
Query: 378 QWPKTAQILSDLGYASWMQQNLTLTQCSH*MKVLGRLCSHASD*GGYHSQVS*PTRFSFT 437
+W +TAQILSD G S MQQN TL QCSH KVL +L S A D G Y S+ +*PTRFSFT
Sbjct: 568 RWLRTAQILSDSGCVSLMQQNPTLIQCSHWTKVLAQLFSLAGDSGAYPSRGN*PTRFSFT 747
Query: 438 LVCMLSSLKCYLLHLLARVTRECSMY*TDAKRFASLRLETVLSETRRF*QT*GSMKQCDP 497
C L SLKC LHLL R TRECSM * DA+ SLRLET L F*+T*GSMKQCDP
Sbjct: 748 *ECTLRSLKCCPLHLLGRATRECSMC*MDARSLGSLRLETALLAMWHF*RT*GSMKQCDP 927
Query: 498 FGCRRAR*L*KHARHWQRRCRG*MWRFSVKVNRQIVTWKMGRESRRCICIVQWLGKGRMH 557
FGCR +* + A QRRCRG*MWR S+K +++IV WKMGR RRCICIV WLGKG+MH
Sbjct: 928 FGCRPVK*PSEPASC*QRRCRG*MWRSSMKTSKKIVVWKMGRR*RRCICIVHWLGKGKMH 1107
Query: 558 QTMYG 562
MYG
Sbjct: 1108LNMYG 1122
Score = 172 bits (437), Expect(2) = e-119
Identities = 91/113 (80%), Positives = 96/113 (84%)
Frame = +3
Query: 203 SRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSI 262
+RSPNLKSL+LNRSVPVDALQRI+ RAPQL DLGIGS HD S+AY K TILK KSI
Sbjct: 21 ARSPNLKSLKLNRSVPVDALQRIMMRAPQLSDLGIGSLVHDPESEAYIKLKNTILKRKSI 200
Query: 263 TSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRL 315
TSLSGFLEVAP LAAIYPIC NLTSLNLSYAAGI G +LIKLIRHC KLQRL
Sbjct: 201 TSLSGFLEVAPHCLAAIYPICPNLTSLNLSYAAGIQGSDLIKLIRHCVKLQRL 359
>TC225843 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein (At3g26830), partial (35%)
Length = 957
Score = 285 bits (730), Expect = 3e-77
Identities = 158/245 (64%), Positives = 175/245 (70%)
Frame = +1
Query: 318 D*VL*LAHVKNCKN*EFSHQLHLETKQLLPKKDL*RYQWDVLSFTHYSTSVTR*PMLLS* 377
D*VL*L HVK CKN* F H L +QL KK +YQW LSFT YSTS TR* ML S*
Sbjct: 1 D*VL*LRHVKICKN*GFFHLFPLGIQQLSQKKGWLQYQWAALSFTRYSTSATR*QMLHS* 180
Query: 378 QWPKTAQILSDLGYASWMQQNLTLTQCSH*MKVLGRLCSHASD*GGYHSQVS*PTRFSFT 437
QW +TAQIL D G S MQQNLTL QCSH KVL +LCS A D G Y S+ +* TRFSFT
Sbjct: 181 QWLRTAQILFDSGCVSLMQQNLTLIQCSHWTKVLAQLCSLAGDSGAYLSRGN*LTRFSFT 360
Query: 438 LVCMLSSLKCYLLHLLARVTRECSMY*TDAKRFASLRLETVLSETRRF*QT*GSMKQCDP 497
CML +LKC LLHLL VTREC M * DA+ SLR ET L T F*+T*GSMKQCDP
Sbjct: 361 *ECMLRNLKCCLLHLLGMVTRECCMC*MDARSLGSLR*ETALLATWHF*RT*GSMKQCDP 540
Query: 498 FGCRRAR*L*KHARHWQRRCRG*MWRFSVKVNRQIVTWKMGRESRRCICIVQWLGKGRMH 557
FGCR +*L + A RRC+G*MWR S+K +++IV WKMGR RRCICIV WLGKG+MH
Sbjct: 541 FGCRPVK*LSEPASC*PRRCQG*MWRSSMKTSKKIVVWKMGRR*RRCICIVHWLGKGKMH 720
Query: 558 QTMYG 562
Q MYG
Sbjct: 721 QNMYG 735
>BE805896 similar to GP|9279671|dbj transport inhibitor response-like protein
{Arabidopsis thaliana}, partial (23%)
Length = 420
Score = 233 bits (593), Expect = 2e-61
Identities = 113/138 (81%), Positives = 129/138 (92%)
Frame = +2
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWL 171
V+D+SLELLSRSF+NFKSLVLVSCEGFTTDGLAA+AANCR L+ELDLQENEV+DH+GQWL
Sbjct: 2 VTDKSLELLSRSFMNFKSLVLVSCEGFTTDGLAAIAANCRFLKELDLQENEVDDHRGQWL 181
Query: 172 SCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQ 231
SCFP+ CTSLVSLNFACLKG INLGALERLV+RSPNLKSLRLN +VP++ALQRIL RAPQ
Sbjct: 182 SCFPDCCTSLVSLNFACLKGQINLGALERLVARSPNLKSLRLNHTVPLNALQRILRRAPQ 361
Query: 232 LMDLGIGSFFHDLNSDAY 249
++DLGIGSF D NS+ +
Sbjct: 362 IVDLGIGSFIPDPNSNVF 415
>BM094881 similar to PIR|T48087|T48 transport inhibitor response protein TIR1
[imported] - Arabidopsis thaliana, partial (23%)
Length = 420
Score = 206 bits (524), Expect = 2e-53
Identities = 92/139 (66%), Positives = 120/139 (86%)
Frame = +1
Query: 27 LVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKSLTLKGKPHFADFSLVPHGWGG 86
LVCKSWY IER+ R+RVF+GNCY++SP +V RFP ++S+ +KGKPHFADF+LVP GWG
Sbjct: 1 LVCKSWYEIERWCRRRVFVGNCYAVSPATVVNRFPKVRSIAIKGKPHFADFNLVPEGWGA 180
Query: 87 FVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAV 146
+V PWI+A+A L+E+RLKRMV++DE LEL+++SF NF+ LVL SCEGFTTDGLAA+
Sbjct: 181 YVGPWIKAMAAAYPWLQEIRLKRMVIADECLELIAKSFKNFQVLVLTSCEGFTTDGLAAI 360
Query: 147 AANCRSLRELDLQENEVED 165
AANCR+LREL+L+E+EV+D
Sbjct: 361 AANCRNLRELELRESEVDD 417
>TC225845 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein (At3g26830), partial (23%)
Length = 672
Score = 181 bits (458), Expect = 1e-45
Identities = 98/163 (60%), Positives = 113/163 (69%)
Frame = +1
Query: 400 TLTQCSH*MKVLGRLCSHASD*GGYHSQVS*PTRFSFTLVCMLSSLKCYLLHLLARVTRE 459
TL QCSH KVL +LCS AS+ GG HS S* +FSFTL CMLSSL+C LLHLL +VTR
Sbjct: 52 TLIQCSHSTKVLAQLCSLASNFGGCHSPAS*LIKFSFTLGCMLSSLRCCLLHLLEKVTRL 231
Query: 460 CSMY*TDAKRFASLRLETVLSETRRF*QT*GSMKQCDPFGCRRAR*L*KHARHWQRRCRG 519
CSM * DA+ FA+LR E L T RF T*GSMK+CDPFGC A + ARH RRC+
Sbjct: 232 CSMC*MDARNFANLR*EIALLVTPRFSWT*GSMKRCDPFGCHPAMSPLEPARHLPRRCQD 411
Query: 520 *MWRFSVKVNRQIVTWKMGRESRRCICIVQWLGKGRMHQTMYG 562
*MWR ++ R IV W M R+ RR ICI WLG+G+ HQ MYG
Sbjct: 412 *MWRSLMETKR*IVMWTMARK*RRRICIEHWLGEGKTHQNMYG 540
>TC221662 weakly similar to UP|Q6TDU2 (Q6TDU2) Coronatine-insensitive 1,
partial (44%)
Length = 993
Score = 158 bits (399), Expect = 7e-39
Identities = 98/291 (33%), Positives = 155/291 (52%), Gaps = 2/291 (0%)
Frame = +2
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D V++ V Y+ DR+++S VC+ Y ++ TR+ V I CY+ +P+RL RFP L+S
Sbjct: 68 DVVLDCVMPYIHDSKDRDAVSQVCRRLYELDSLTRKHVTIALCYTTTPDRLRRRFPHLES 247
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
L LKGKP A F+L+P WGGFV PW+ +++ L+ L +RM+V D L++L+RS
Sbjct: 248 LNLKGKPRAAMFNLIPEDWGGFVTPWVREISQYFDCLKSLHFRRMIVRDSDLQVLARSRG 427
Query: 126 N-FKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSL 184
+ ++L L C GF+TDGL + CR+LR L L+E+ + ++ G WL + T L +L
Sbjct: 428 HILQALKLDKCSGFSTDGLYYIGRYCRNLRVLFLEESSLVENDGDWLHELALNNTVLETL 607
Query: 185 NFACLK-GDINLGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHD 243
NF ++ + LE + PNL S+++ +D L A L + GS+
Sbjct: 608 NFYLTDIANVRIQDLELIARNCPNLNSVKITDCEVLD-LVNFFRYASALEEFCGGSY--- 775
Query: 244 LNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYA 294
++ + A L K S G + + ++P L L+L YA
Sbjct: 776 --NEESEKYSAISLPAK--LSRLGLTYITKNEMPMVFPYAALLKKLDLLYA 916
>BM521565 similar to PIR|T48087|T48 transport inhibitor response protein TIR1
[imported] - Arabidopsis thaliana, partial (23%)
Length = 428
Score = 105 bits (261), Expect(2) = 2e-36
Identities = 52/72 (72%), Positives = 60/72 (83%), Gaps = 1/72 (1%)
Frame = +2
Query: 124 FVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEV-EDHKGQWLSCFPESCTSLV 182
F NFK LVL SCEGFTTDGLAA+AANCR+LRELDLQE+EV ED G WLS FP+S TSLV
Sbjct: 2 FKNFKVLVLTSCEGFTTDGLAAIAANCRNLRELDLQESEVEEDLSGHWLSHFPDSYTSLV 181
Query: 183 SLNFACLKGDIN 194
SLN +CL +++
Sbjct: 182 SLNISCLNNEVS 217
Score = 66.2 bits (160), Expect(2) = 2e-36
Identities = 30/70 (42%), Positives = 47/70 (66%)
Frame = +3
Query: 195 LGALERLVSRSPNLKSLRLNRSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKA 254
L ALERL+ R PNL++LRLNR+VP+D L +L + PQL++LG G + ++ + ++ +A
Sbjct: 219 LSALERLLGRCPNLRTLRLNRAVPLDRLPNLLLQCPQLVELGTGVYSTEMRPEVFSNLEA 398
Query: 255 TILKCKSITS 264
CK + S
Sbjct: 399 AFSGCKQLKS 428
>TC234386 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (14%)
Length = 467
Score = 129 bits (324), Expect = 3e-30
Identities = 52/82 (63%), Positives = 72/82 (87%)
Frame = +1
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP+EV+EHVF ++ + DRN++SLVCKSWY IER+ R++VF+GNCY++SP +V+RFP++
Sbjct: 220 FPEEVLEHVFSFIWNERDRNAISLVCKSWYEIERWCRRKVFVGNCYAVSPLMVVKRFPEV 399
Query: 64 KSLTLKGKPHFADFSLVPHGWG 85
+S+ LKGKPHFADF+LVP GWG
Sbjct: 400 RSIALKGKPHFADFNLVPDGWG 465
>BM732420 weakly similar to GP|13249030|gb F-box containing protein TIR1
{Populus tremula x Populus tremuloides}, partial (19%)
Length = 425
Score = 127 bits (318), Expect = 2e-29
Identities = 73/135 (54%), Positives = 93/135 (68%), Gaps = 4/135 (2%)
Frame = +3
Query: 102 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQE- 160
L +L LKRM ++D L LLS S +F+ L+L CEGF T LAA+A+NCR LR L+L E
Sbjct: 21 LNKLHLKRMSLTDHDLTLLSHSLPSFQDLLLTCCEGFGTTALAALASNCRLLRVLELVEC 200
Query: 161 -NEVEDHKGQWLSCFPE--SCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSV 217
EV D + W+SCFPE + T L SL F C++ INL ALERLV+RSP+L+ LRLNR V
Sbjct: 201 VVEVGDEELDWISCFPEIDAQTYLESLVFDCVECPINLEALERLVARSPSLRKLRLNRYV 380
Query: 218 PVDALQRILTRAPQL 232
+ L R++ RAPQL
Sbjct: 381 SMSQLHRLMHRAPQL 425
>TC225841 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein (At3g26830), partial (10%)
Length = 848
Score = 121 bits (304), Expect = 7e-28
Identities = 54/60 (90%), Positives = 59/60 (98%)
Frame = +2
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEH+FDYVVSHSDRN+LSLVCKSWYRIER TRQRVFIGNCYSI+PERL++RF
Sbjct: 668 MNYFPDEVIEHIFDYVVSHSDRNALSLVCKSWYRIERCTRQRVFIGNCYSITPERLIQRF 847
>TC234752 weakly similar to UP|Q6TDU2 (Q6TDU2) Coronatine-insensitive 1,
partial (25%)
Length = 502
Score = 117 bits (292), Expect = 2e-26
Identities = 62/152 (40%), Positives = 91/152 (59%), Gaps = 1/152 (0%)
Frame = +2
Query: 6 DEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKS 65
D V++ V Y+ DR+++S VC+ WY ++ TR+ V I CY+ +P RL RFP L+S
Sbjct: 47 DVVLDCVIPYIDDPKDRDAVSQVCRRWYELDSLTRKHVTIALCYTTTPARLRRRFPHLES 226
Query: 66 LTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRSFV 125
L LKGKP A F+L+P WGG V PW++ +++ L+ L +RM+V D L+ L+R
Sbjct: 227 LKLKGKPRAAMFNLIPEDWGGHVTPWVKEISQYFDCLKSLHFRRMIVKDSDLQNLARDRG 406
Query: 126 N-FKSLVLVSCEGFTTDGLAAVAANCRSLREL 156
+ +L L TTDGL + C SLR+L
Sbjct: 407 HMLHALKLD*SS*STTDGLFHICRFCISLRDL 502
>AW755995 similar to PIR|T48087|T480 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana, partial (12%)
Length = 292
Score = 112 bits (279), Expect = 6e-25
Identities = 47/89 (52%), Positives = 67/89 (74%)
Frame = +2
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP+EV+EHVF ++ S DRN++SLVCKSWY I+R+ R++VF GNCY++SP ++RFP+L
Sbjct: 26 FPEEVLEHVFSFIWSERDRNAISLVCKSWYEIDRWCRRKVFEGNCYAVSPLMGIKRFPEL 205
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWI 92
KS+ L KPH AD L+ + G + Y W+
Sbjct: 206 KSIVLIRKPHLADLHLITNSGGEYGYTWL 292
>TC228955 weakly similar to PIR|T48087|T48087 transport inhibitor response
protein TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (43%)
Length = 1480
Score = 75.5 bits (184), Expect = 6e-14
Identities = 41/100 (41%), Positives = 57/100 (57%)
Frame = +3
Query: 217 VPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSL 276
V ++ LQR++ PQL +LG GSF +L S + ++ + CK++ +LSG L
Sbjct: 54 VTLEQLQRLIVHVPQLGELGTGSFSQELTSQQCSDLESALKNCKNLHTLSGLWVATAQYL 233
Query: 277 AAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRLW 316
+Y C NLT LN SYA + L KL+ HC KLQRLW
Sbjct: 234 PVLYSACTNLTFLNFSYAP-LDSDGLTKLLVHCPKLQRLW 350
Score = 38.1 bits (87), Expect = 0.011
Identities = 51/165 (30%), Positives = 66/165 (39%)
Frame = +1
Query: 357 DVLSFTHYSTSVTR*PMLLS*QWPKTAQILSDLGYASWMQQNLTLTQCSH*MKVLGRLCS 416
D FT + T +*PMLL KT L AS + + M+ L L
Sbjct: 499 DAQDFTMFYTFAVK*PMLLLLLLSKTVLTLLISDSASCTLVS*IT*PKNQWMRPLELLLR 678
Query: 417 HASD*GGYHSQVS*PTRFSFTLVCMLSSLKCYLLHLLARVTRECSMY*TDAKRFASLRLE 476
A + G QV * T M + K + LL V C++ T + + R
Sbjct: 679 LALNSRGLQYQVI*LT*LLSI*ASMQKTWKLFPWLLLEAVIGACAVCWTAVQS*EN*R*G 858
Query: 477 TVLSETRRF*QT*GSMKQCDPFGCRRAR*L*KHARHWQRRCRG*M 521
TV T F Q GSM Q D GC+ A * * W ++C G*M
Sbjct: 859 TVHLATELFCQAWGSMSQ*DHCGCQTAI*Q*MVLDCWPKKCLG*M 993
Score = 30.4 bits (67), Expect = 2.2
Identities = 35/145 (24%), Positives = 63/145 (43%), Gaps = 7/145 (4%)
Frame = +3
Query: 100 VGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTD-------GLAAVAANCRS 152
VG RL R+ + +LE L R V+ L + F+ + L + NC++
Sbjct: 15 VGANH*RL*RL--TKVTLEQLQRLIVHVPQLGELGTGSFSQELTSQQCSDLESALKNCKN 188
Query: 153 LRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLR 212
L L + + Q+L +CT+L LNF+ ++ L +L+ P L+ L
Sbjct: 189 LHTL----SGLWVATAQYLPVLYSACTNLTFLNFS--YAPLDSDGLTKLLVHCPKLQRLW 350
Query: 213 LNRSVPVDALQRILTRAPQLMDLGI 237
+ +V L+ + + P L +L +
Sbjct: 351 VVDTVEDKGLEAVGSHCPLLEELRV 425
>TC221974 similar to UP|Q9AUH6 (Q9AUH6) F-box containing protein TIR1,
partial (25%)
Length = 611
Score = 68.6 bits (166), Expect = 7e-12
Identities = 43/92 (46%), Positives = 50/92 (53%)
Frame = +2
Query: 225 ILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPICQ 284
++ RAP G GSF D F + CKS+ LSGF E+ P L AIYP C
Sbjct: 2 LMHRAPS*XS-GTGSFSAS-ELDQELDFASAFAACKSLVCLSGFREIWPDYLPAIYPACA 175
Query: 285 NLTSLNLSYAAGILGIELIKLIRHCGKLQRLW 316
NL SLN SY A I +LI +IRHC KLQ W
Sbjct: 176 NLISLNFSY-ADISADQLISVIRHCHKLQTFW 268
>TC223250 similar to UP|Q6TDU2 (Q6TDU2) Coronatine-insensitive 1, partial
(15%)
Length = 481
Score = 56.2 bits (134), Expect = 4e-08
Identities = 36/111 (32%), Positives = 55/111 (49%), Gaps = 1/111 (0%)
Frame = +2
Query: 50 SISPERLVERFPDLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKR 109
S+S ++V+ SL A FSL+P WG V PW++ +++ L+ L +R
Sbjct: 29 SLSLSKMVDDPMLRSSLPAPRVAQAAMFSLIPEDWGEHVSPWVKEISQYFDCLKSLHFRR 208
Query: 110 MVVSDESLELLSRSFVN-FKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQ 159
M+V D L+ L+R + +L L C FTTDGL + C L L+
Sbjct: 209 MIVKDSDLQNLARDRGHVLHALKLDKCFSFTTDGLFHIGRFCNDKLGLSLE 361
>BE804193 similar to GP|13249030|gb| F-box containing protein TIR1 {Populus
tremula x Populus tremuloides}, partial (9%)
Length = 231
Score = 56.2 bits (134), Expect = 4e-08
Identities = 30/60 (50%), Positives = 38/60 (63%)
Frame = +1
Query: 252 FKATILKCKSITSLSGFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGK 311
+ A C+S+ LSGF E+ L AIYP+C NLTSLNLSY A + +L +IRHC K
Sbjct: 55 YAAAFEACRSLVCLSGFREIWADYLPAIYPVCTNLTSLNLSY-ADVNTDQLKSVIRHCHK 231
>TC219384
Length = 1313
Score = 51.2 bits (121), Expect = 1e-06
Identities = 55/204 (26%), Positives = 87/204 (41%), Gaps = 4/204 (1%)
Frame = +2
Query: 116 SLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFP 175
S+ +S S SL + SC ++ + C + ELDL +NE++D +S
Sbjct: 8 SIASISNSCAGLTSLKMESCTLVPSEAFVLIGEKCHYIEELDLTDNEIDDEGLMSIS--- 178
Query: 176 ESCTSLVSLNFA-CLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQ-RILTRAPQLM 233
SC+ L SL CL +I L + LK L L RS VD L + R +
Sbjct: 179 -SCSRLSSLKIGICL--NITDRGLTYVGMHCSKLKELDLYRSTGVDDLGISAIARGCPGL 349
Query: 234 DLGIGSFFHDLNSDAYAMFKATILKCKSITSLS--GFLEVAPFSLAAIYPICQNLTSLNL 291
++ S+ + A T+ KC ++ +L G L V LAAI C+ L+ L++
Sbjct: 350 EMINTSYCTSITDRAL----ITLSKCSNLKTLEIRGCLLVTSIGLAAIAMNCRQLSRLDI 517
Query: 292 SYAAGILGIELIKLIRHCGKLQRL 315
I +I L L+++
Sbjct: 518 KKCYNIDDSGMIALAHFSQNLRQI 589
Score = 47.0 bits (110), Expect = 2e-05
Identities = 54/210 (25%), Positives = 89/210 (41%), Gaps = 4/210 (1%)
Frame = +2
Query: 102 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQEN 161
+EEL L + DE L +S S SL + C T GL V +C L+ELDL +
Sbjct: 119 IEELDLTDNEIDDEGLMSIS-SCSRLSSLKIGICLNITDRGLTYVGMHCSKLKELDLYRS 295
Query: 162 EVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDA 221
D G +S C L +N + + + +S+ NLK+L + + V +
Sbjct: 296 TGVDDLG--ISAIARGCPGLEMINTSYCTSITDRALIT--LSKCSNLKTLEIRGCLLVTS 463
Query: 222 LQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYP 281
+ G+ + + + + + I KC +I SG + +A FS
Sbjct: 464 I-------------GLAA----IAMNCRQLSRLDIKKCYNIDD-SGMIALAHFS------ 571
Query: 282 ICQNLTSLNLSYAA----GILGIELIKLIR 307
QNL +NLSY++ G+L + I ++
Sbjct: 572 --QNLRQINLSYSSVTDVGLLSLANISCLQ 655
Score = 40.8 bits (94), Expect = 0.002
Identities = 26/70 (37%), Positives = 40/70 (57%), Gaps = 1/70 (1%)
Frame = +2
Query: 92 IEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANC 150
I A+A+ GLE + ++D +L LS+ N K+L + C T+ GLAA+A NC
Sbjct: 317 ISAIARGCPGLEMINTSYCTSITDRALITLSKCS-NLKTLEIRGCLLVTSIGLAAIAMNC 493
Query: 151 RSLRELDLQE 160
R L LD+++
Sbjct: 494 RQLSRLDIKK 523
>TC231852 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (30%)
Length = 542
Score = 48.5 bits (114), Expect = 8e-06
Identities = 25/39 (64%), Positives = 27/39 (69%)
Frame = +1
Query: 278 AIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRLW 316
A+YPIC LTSLNLSYA I +LIKLI C L RLW
Sbjct: 4 AVYPICSRLTSLNLSYAI-IQSSDLIKLISQCPNLLRLW 117
Score = 29.3 bits (64), Expect = 4.9
Identities = 25/80 (31%), Positives = 36/80 (44%), Gaps = 8/80 (10%)
Frame = +1
Query: 142 GLAAVAANCRSLREL--------DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDI 193
GL A+AA+C+ LREL L+ N +G L E C L S+ + C + +
Sbjct: 142 GLYALAASCKDLRELRVFPSDPFGLEPNVALTEQG--LVSVSEGCPKLQSVLYFCRQ--M 309
Query: 194 NLGALERLVSRSPNLKSLRL 213
+ AL + NL RL
Sbjct: 310 SNAALHTIARNRTNLTRFRL 369
>TC217926 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (20%)
Length = 608
Score = 47.8 bits (112), Expect = 1e-05
Identities = 32/86 (37%), Positives = 42/86 (48%)
Frame = +3
Query: 447 CYLLHLLARVTRECSMY*TDAKRFASLRLETVLSETRRF*QT*GSMKQCDPFGCRRAR*L 506
CYL LL V + M S R T TR F Q S +QCDPFGC AR*+
Sbjct: 3 CYLWLLLXIVIWDFIMCYQGVIILGSWRSGTAPLVTRPFWQMLKSWRQCDPFGCPPAR*V 182
Query: 507 *KHARHWQRRCRG*MWRFSVKVNRQI 532
+H +W R+C+ + +K + QI
Sbjct: 183 MEHVSYWVRKCQDSTLKSLMKEDLQI 260
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.340 0.146 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,926,199
Number of Sequences: 63676
Number of extensions: 448313
Number of successful extensions: 3436
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 3373
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3421
length of query: 562
length of database: 12,639,632
effective HSP length: 102
effective length of query: 460
effective length of database: 6,144,680
effective search space: 2826552800
effective search space used: 2826552800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC133780.9


