

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.6 - phase: 0
(1061 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
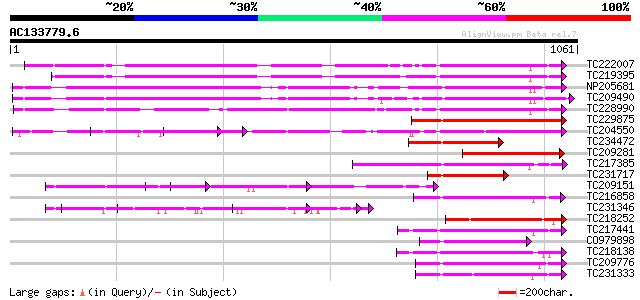
Score E
Sequences producing significant alignments: (bits) Value
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 502 e-142
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 468 e-132
NP205681 receptor protein kinase-like protein 465 e-131
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 459 e-129
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 453 e-127
TC229875 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repea... 384 e-106
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 284 2e-76
TC234472 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repea... 262 5e-70
TC209281 similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat recep... 238 1e-62
TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase P... 226 5e-59
TC231717 weakly similar to UP|Q8VZG8 (Q8VZG8) AT4g08850/T32A17_1... 199 5e-51
TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-l... 199 7e-51
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 197 3e-50
TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 192 5e-49
TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial ... 189 4e-48
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 174 2e-43
CO979898 174 2e-43
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 171 1e-42
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 170 3e-42
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 169 5e-42
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 502 bits (1292), Expect = e-142
Identities = 352/1025 (34%), Positives = 518/1025 (50%), Gaps = 11/1025 (1%)
Frame = +3
Query: 28 ALLKWKASFDNQSQSILSTWKNTTNPCSKWRGIECDKSNLISTIDLANLGLKGTLHSLTF 87
ALL ++ + + +LS+W N + P W G+ CD ++ ++L L L GTL S
Sbjct: 105 ALLSLRSVITDATPPVLSSW-NASIPYCSWLGVTCDNRRHVTALNLTGLDLSGTL-SADV 278
Query: 88 SSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDFF 147
+ P L L++ N F G IPP + LS + LN S N + P E++ L+SL+ LD +
Sbjct: 279 AHLPFLSNLSLAANKFSGPIPPSLSALSGLRYLNLSNNVFNETFPSELWRLQSLEVLDLY 458
Query: 148 FCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQE 207
++G + ++ + NL +L LGGN FSG IPPE G+ ++L+
Sbjct: 459 NNNMTGVLPLAVAQMQNLRHLHLGGNFFSG-QIPPEYGRWQRLQ---------------- 587
Query: 208 IGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIY 267
Y+ +S N L G IP IGN++ L +L G IP + N+S L +
Sbjct: 588 --------YLAVSGNELDGTIPPEIGNLTSLRELYIGYYNTYTGGIPPEIGNLSELVRLD 743
Query: 268 LYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPA 327
+ +LSG IP ++ L LD L L +N LSG + +GNLK+L + L NN LSG IPA
Sbjct: 744 VAYCALSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPA 923
Query: 328 SIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFV 387
S G L N+ ++ N L G IP IG L L V ++ N L G IP GL
Sbjct: 924 SFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLTGSIPEGLGKNGRLNLVD 1103
Query: 388 VSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAE 447
+S N G LP +C+G +L+ L N GP+P SL +C S+ RIR+ N + G I +
Sbjct: 1104 LSSNKLTGTLPPYLCSGNTLQTLITLGNFLFGPIPESLGTCESLTRIRMGENFLNGSIPK 1283
Query: 448 DFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLH 507
P L V+L DN +SG P LG++
Sbjct: 1284 GLFGLPKLTQVELQDNY------------------------LSGEFPEVGSVAVNLGQIT 1391
Query: 508 LSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIP 567
LS+NQL+G L I G S+ L + N FT IPT+IG LQ+L ++D GN+ SG I
Sbjct: 1392 LSNNQLSGALSPSI-GNFSSVQKLLLDGNMFTGRIPTQIGRLQQLSKIDFSGNKFSGPIA 1568
Query: 568 NEVAELPKLRMLNLSRNRIEGRIPSTFDSALASIDLSGNRLNGNIPTSLGFLVQLSMLNL 627
E+++ L L+LSRN L+G+IP + + L+ LNL
Sbjct: 1569 PEISQCKLLTFLDLSRNE----------------------LSGDIPNEITGMRILNYLNL 1682
Query: 628 SHNMLSGTIPSTFS--MSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGL 685
S N L G+IPS+ S SL V+ S N L G +P F + SF N LCG G
Sbjct: 1683 SKNHLVGSIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFLGNPDLCGPYLGA 1862
Query: 686 VPCATSQ-IHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQK 744
+ H K + S+ + L + L+L + ++ F+ + ++ E +
Sbjct: 1863 CKGGVANGAHQPHVKGLSSSLKLLL-VVGLLLCSIAFAVAAIFKARS----LKKASEARA 2027
Query: 745 GVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRD 804
L + D ++++ + + +IG G G VYK +P G VAVK+L
Sbjct: 2028 WKLTAFQRLD--FTVDDVLHCLK---EDNIIGKGGAGIVYKGAMPNGDHVAVKRL----- 2177
Query: 805 EEMSFFSSKS--FTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNE 862
MS SS F +EI+TL I+HR+I++L GFCS+ + + LVY++M GSL ++L+ +
Sbjct: 2178 PAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGK 2357
Query: 863 KQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAK 922
K W+ R + A L YLHHDCSP I+HRD+ S NILL+ ++EAHV+DFG AK
Sbjct: 2358 KGG-HLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAK 2534
Query: 923 FLKPDLHS--WTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP----GDL 976
FL+ S + AG++GY APE + T++V+EK DVYSFGV+ LE+I G+ P GD
Sbjct: 2535 FLQDSGTSECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVGEFGDG 2714
Query: 977 ISLFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQ 1036
+ + +N + +VLD R V P+ EV+ + +A C+ + RPTM +
Sbjct: 2715 VDIVQWVRKMTDSNKEGVLKVLDPRLPSV--PL-HEVMHVFYVAMLCVEEQAVERPTMRE 2885
Query: 1037 VCKML 1041
V ++L
Sbjct: 2886 VVQIL 2900
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 468 bits (1205), Expect = e-132
Identities = 335/975 (34%), Positives = 479/975 (48%), Gaps = 11/975 (1%)
Frame = +3
Query: 78 LKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYT 137
L G L S + P L L++ +N F G IPP + LS + LN S N + P E+
Sbjct: 3 LSGPL-SADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSR 179
Query: 138 LRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQ 197
L++L+ LD + ++G + ++ + NL +L LGGN FSG IPPE G+ ++L+
Sbjct: 180 LQNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSG-QIPPEYGRWQRLQ------ 338
Query: 198 GSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSL 257
Y+ +S N L G IP IGN+S L +L G IP +
Sbjct: 339 ------------------YLAVSGNELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEI 464
Query: 258 WNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLR 317
N+S L + LSG IP ++ L LD L L +N LSG + +GNLK+L + L
Sbjct: 465 GNLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLS 644
Query: 318 NNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGL 377
NN LSG IPA G L N+ ++ N L G IP IG L L V ++ N G IP GL
Sbjct: 645 NNMLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGL 824
Query: 378 YNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIE 437
+S N G LP+ +C+G +L+ L N GP+P SL SC S+ RIR+
Sbjct: 825 GKNGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMG 1004
Query: 438 GNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDF 497
N + G I P L V+L DN +SG P
Sbjct: 1005 ENFLNGSIPRGLFGLPKLTQVELQDNY------------------------LSGEFPEVG 1112
Query: 498 IGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDL 557
LG++ LS+NQL+G LP I G S+ L + N FT IP +IG LQ+L ++D
Sbjct: 1113 SVAVNLGQITLSNNQLSGVLPPSI-GNFSSVQKLILDGNMFTGRIPPQIGRLQQLSKIDF 1289
Query: 558 GGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSALASIDLSGNRLNGNIPTSLG 617
GN+ SG I E+++ L L+LSRN L+G+IP +
Sbjct: 1290 SGNKFSGPIVPEISQCKLLTFLDLSRNE----------------------LSGDIPNEIT 1403
Query: 618 FLVQLSMLNLSHNMLSGTIPSTFS--MSLDFVNISDNQLDGPLPENPAFLRAPFESFKNN 675
+ L+ LNLS N L G IPS+ S SL V+ S N L G +P F + SF N
Sbjct: 1404 GMRILNYLNLSRNHLVGGIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFLGN 1583
Query: 676 KGLCGNITGLVPCATSQIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNEE 735
LCG G + + L S F L + L+L + ++ F+ +
Sbjct: 1584 PDLCGPYLGACKDGVANGAHQPHVKGLSSSFKLLLVVGLLLCSIAFAVAAIFKARS---- 1751
Query: 736 IQTEEEVQKGVLFSIWSHDGKMMFE-NIIEATENFDDKYLIGVGSQGNVYKAELPTGLVV 794
++K W + + + + +IG G G VYK +P G V
Sbjct: 1752 ------LKKASGARAWKLTAFQRLDFTVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHV 1913
Query: 795 AVKKLHLVRDEEMSFFSSKS--FTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEG 852
AVK+L MS SS F +EI+TL I+HR+I++L GFCS+ + + LVY++M
Sbjct: 1914 AVKRL-----PAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPN 2078
Query: 853 GSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYE 912
GSL ++L+ +K W+ R + A L YLHHDCSP I+HRD+ S NILL+ ++E
Sbjct: 2079 GSLGEVLHGKKGG-HLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHE 2255
Query: 913 AHVSDFGTAKFLKPDLHS--WTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIG 970
AHV+DFG AKFL+ S + AG++GY APE + T++V+EK DVYSFGV+ LE+I G
Sbjct: 2256 AHVADFGLAKFLQDSGTSECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITG 2435
Query: 971 KHP----GDLISLFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQ 1026
+ P GD + + +N + +VLD R V P+ EV+ + +A C+ +
Sbjct: 2436 RKPVGEFGDGVDIVQWVRKMTDSNKEGVLKVLDPRLPSV--PL-HEVMHVFYVAMLCVEE 2606
Query: 1027 VPRSRPTMDQVCKML 1041
RPTM +V ++L
Sbjct: 2607 QAVERPTMREVVQIL 2651
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 465 bits (1197), Expect = e-131
Identities = 331/1056 (31%), Positives = 515/1056 (48%), Gaps = 20/1056 (1%)
Frame = +1
Query: 6 FIIMILCVLPTLSVAEDSEAKLALLKWKASF--DNQSQSILSTWKNTTNPCSK--WRGIE 61
F+ I + T S D +A LLK K S D L WK +T+ + + G+
Sbjct: 34 FVFFIWLHVATCSSFSDMDA---LLKLKESMKGDRAKDDALHDWKFSTSLSAHCFFSGVS 204
Query: 62 CDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLN 121
CD+ + I+++ + L +G +PP+IG L ++ L
Sbjct: 205 CDQELRVVAINVSFVPL-------------------------FGHVPPEIGELDKLENLT 309
Query: 122 FSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEID-KSIGNLTNLSYLDLGGNNFSGGPI 180
S+N + G +P+E+ L SLK L+ SG K I +T L LD+ NNF+G +
Sbjct: 310 ISQNNLTGELPKELAALTSLKHLNISHNVFSGYFPGKIILPMTELEVLDVYDNNFTGS-L 486
Query: 181 PPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQ 240
P E KL+KL+YL + GSIP+ +L ++ LS N LSG IP+++ + L
Sbjct: 487 PEEFVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGNIPKSLSKLKTLRI 666
Query: 241 LMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGF 300
L N G IP M SL + L + +LSG IP S+ N+ NLD L L MNNL+G
Sbjct: 667 LKLGYNNAYEGGIPPEFGTMESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLTGT 846
Query: 301 IPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLI 360
IPS + ++ +L L L N L+G IP L NL + NNL G++P+ +G L L
Sbjct: 847 IPSELSDMVSLMSLDLSFNGLTGEIPTRFSQLKNLTLMNFFHNNLRGSVPSFVGELPNLE 1026
Query: 361 VFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGP 420
++ N +P L + F V++N F G +P +C G L+ N F GP
Sbjct: 1027 TLQLWENNFSSELPQNLGQNGKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGP 1206
Query: 421 VPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLE 480
+P + +C S+ +IR N + G + P++ ++L++N+F+G + P
Sbjct: 1207 IPNEIANCKSLTKIRASNNYLNGAVPSGIFKLPSVTIIELANNRFNGELPP--------- 1359
Query: 481 TFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTD 540
ISG LG L LS+N TGK+P L +++L L + N F
Sbjct: 1360 -------EISG---------DSLGILTLSNNLFTGKIP-PALKNLRALQTLSLDTNEFLG 1488
Query: 541 SIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSA--L 598
IP E+ L L +++ GN L+G IP L ++LSRN ++G IP + L
Sbjct: 1489 EIPGEVFDLPMLTVVNISGNNLTGPIPTTFTRCVSLAAVDLSRNMLDGEIPKGMKNLTDL 1668
Query: 599 ASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMSLDFVNISDNQLDGPL 658
+ ++S N+++G++P + F++ L+ L+LS+N G +P+ F+ SD
Sbjct: 1669 SIFNVSINQISGSVPDEIRFMLSLTTLDLSYNNFIGKVPT----GGQFLVFSD------- 1815
Query: 659 PENPAFLRAPFESFKNNKGLCGNITGLVPCATSQIHSRKSKNILQSVFIALGALILVLSG 718
+SF N LC + + C S + R+ L+S + + + L +
Sbjct: 1816 -----------KSFAGNPNLCSSHS----CPNSSLKKRRGPWSLKSTRVIVMVIALATAA 1950
Query: 719 VGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDG-KMMFENIIEATENFDDKYLIGV 777
+ ++ + RR++ K L W G + + E E ++ +IG
Sbjct: 1951 ILVAGTEYMRRRR------------KLKLAMTWKLTGFQRLNLKAEEVVECLKEENIIGK 2094
Query: 778 GSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFC 837
G G VY+ + G VA+K+L + F +EIET+ I+HRNI++L G+
Sbjct: 2095 GGAGIVYRGSMRNGSDVAIKRLVGAGSGRNDY----GFKAEIETVGKIRHRNIMRLLGYV 2262
Query: 838 SHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIH 897
S+ + + L+Y++M GSL + L+ K WE R + A L YLHHDCSP IIH
Sbjct: 2263 SNKETNLLLYEYMPNGSLGEWLHGAKGG-HLKWEMRYKIAVEAAKGLCYLHHDCSPLIIH 2439
Query: 898 RDISSKNILLNLDYEAHVSDFGTAKFLKP--DLHSWTQFAGTFGYAAPELSQTMEVNEKC 955
RD+ S NILL+ +EAHV+DFG AKFL S + AG++GY APE + T++V+EK
Sbjct: 2440 RDVKSNNILLDAHFEAHVADFGLAKFLYDLGSSQSMSSIAGSYGYIAPEYAYTLKVDEKS 2619
Query: 956 DVYSFGVLALEIIIGKHP----GDLISLF------LSPSTRPTANDMLLTEVLDQRPQKV 1005
DVYSFGV+ LE+IIG+ P GD + + ++P+ ++L V P+
Sbjct: 2620 DVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTRLELSQPSDAAVVLAVV---DPRLS 2790
Query: 1006 IKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
P+ VI + +A C+ +V +RPTM +V ML
Sbjct: 2791 GYPL-ISVIYMFNIAMMCVKEVGPTRPTMREVVHML 2895
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 459 bits (1180), Expect = e-129
Identities = 341/1082 (31%), Positives = 526/1082 (48%), Gaps = 30/1082 (2%)
Frame = +1
Query: 6 FIIMILCVLPTLSVAEDSEAKLALLKWKASF--DNQSQSILSTWKNTTNPCSK--WRGIE 61
FI I + T S D E+ LLK K S D L WK + + + G++
Sbjct: 124 FIFFIWLRVATCSSFTDMES---LLKLKDSMKGDKAKDDALHDWKFFPSLSAHCFFSGVK 294
Query: 62 CDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLN 121
CD+ + I+++ + L +G +PP+IG L ++ L
Sbjct: 295 CDRELRVVAINVSFVPL-------------------------FGHLPPEIGQLDKLENLT 399
Query: 122 FSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEID-KSIGNLTNLSYLDLGGNNFSGGPI 180
S+N + G +P+E+ L SLK L+ SG + I +T L LD+ NNF+G P+
Sbjct: 400 VSQNNLTGVLPKELAALTSLKHLNISHNVFSGHFPGQIILPMTKLEVLDVYDNNFTG-PL 576
Query: 181 PPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQ 240
P E+ KL+KL+YL + GSIP+ +L ++ LS N LSG IP+++ + L
Sbjct: 577 PVELVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGKIPKSLSKLKTLRY 756
Query: 241 LMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGF 300
L N G IP +M SL + L + +LSG IP S+ NL NLD L L +NNL+G
Sbjct: 757 LKLGYNNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLTGT 936
Query: 301 IPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLI 360
IPS + + +L L L N L+G IP S L NL + NNL G++P+ +G L L
Sbjct: 937 IPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLE 1116
Query: 361 VFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGP 420
++ N +P L F V +N F G +P +C G L+ + N F GP
Sbjct: 1117 TLQLWDNNFSFVLPPNLGQNGKLKFFDVIKNHFTGLIPRDLCKSGRLQTIMITDNFFRGP 1296
Query: 421 VPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLE 480
+P + +C S+ +IR N + G + P++ ++L++N+F+G + P
Sbjct: 1297 IPNEIGNCKSLTKIRASNNYLNGVVPSGIFKLPSVTIIELANNRFNGELPP--------- 1449
Query: 481 TFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTD 540
ISG LG L LS+N +GK+P L +++L L + N F
Sbjct: 1450 -------EISG---------ESLGILTLSNNLFSGKIP-PALKNLRALQTLSLDANEFVG 1578
Query: 541 SIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSA--L 598
IP E+ L L +++ GN L+G IP + L ++LSRN +EG+IP + L
Sbjct: 1579 EIPGEVFDLPMLTVVNISGNNLTGPIPTTLTRCVSLTAVDLSRNMLEGKIPKGIKNLTDL 1758
Query: 599 ASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMSLDFVNISDNQLDGPL 658
+ ++S N+++G +P + F++ L+ L+LS+N G +P+ F S+
Sbjct: 1759 SIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPT----GGQFAVFSE------- 1905
Query: 659 PENPAFLRAPFESFKNNKGLCGNITGLVPCATSQIH------------SRKSKNILQSVF 706
+SF N LC + + C S ++ S KS ++ +
Sbjct: 1906 -----------KSFAGNPNLCTSHS----CPNSSLYPDDALKKRRGPWSLKSTRVI-VIV 2037
Query: 707 IALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFENIIEAT 766
IALG L+ V +++Y+ RRK + Q+ E+++E
Sbjct: 2038 IALGTAALL---VAVTVYMMRRRKMNLAKTWKLTAFQR----------LNFKAEDVVECL 2178
Query: 767 ENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIK 826
+ ++ +IG G G VY+ +P G VA+K+L + F +EIETL I+
Sbjct: 2179 K---EENIIGKGGAGIVYRGSMPNGTDVAIKRLVGAGSGRNDY----GFKAEIETLGKIR 2337
Query: 827 HRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSY 886
HRNI++L G+ S+ + + L+Y++M GSL + L+ K WE R + A L Y
Sbjct: 2338 HRNIMRLLGYVSNKETNLLLYEYMPNGSLGEWLHGAKGG-HLKWEMRYKIAVEAAKGLCY 2514
Query: 887 LHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD--LHSWTQFAGTFGYAAPE 944
LHHDCSP IIHRD+ S NILL+ D EAHV+DFG AKFL S + AG++GY APE
Sbjct: 2515 LHHDCSPLIIHRDVKSNNILLDGDLEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPE 2694
Query: 945 LSQTMEVNEKCDVYSFGVLALEIIIGKHP----GDLISLF-----LSPSTRPTANDMLLT 995
+ T++V+EK DVYSFGV+ LE+IIG+ P GD + + ++ L+
Sbjct: 2695 YAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTRLELAQPSDAALVL 2874
Query: 996 EVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKMLGAGKSPLENQFHTI 1055
V+D P+ P+ VI + +A C+ ++ +RPTM +V ML P + HT
Sbjct: 2875 AVVD--PRLSGYPL-TSVIYMFNIAMMCVKEMGPARPTMREVVHML---SEPPHSATHTH 3036
Query: 1056 KL 1057
L
Sbjct: 3037 NL 3042
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 453 bits (1165), Expect = e-127
Identities = 331/1049 (31%), Positives = 511/1049 (48%), Gaps = 14/1049 (1%)
Frame = +1
Query: 7 IIMILCVLPTLSVAEDSEAKLALLKWKAS-FDNQSQSILSTWKNTTNPCSKWRGIECDKS 65
+++ L +L A SE + ALL +KAS + LS+W ++T CS W G+ CD
Sbjct: 109 LVLFFLFLHSLQAARISEYR-ALLSFKASSLTDDPTHALSSWNSSTPFCS-WFGLTCDSR 282
Query: 66 NLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKN 125
+++++L +L L GTL + +L ++ L+ + N
Sbjct: 283 RHVTSLNLTSLSLSGTLSD-------------------------DLSHLPFLSHLSLADN 387
Query: 126 PIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIG 185
G IP L +L+ L+ + + L NL LDL NN +G +P +
Sbjct: 388 KFSGPIPASFSALSALRFLNLSNNVFNATFPSQLNRLANLEVLDLYNNNMTG-ELPLSVA 564
Query: 186 KLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFAN 245
+ LR+L + G IP E G +L Y+ LS N L+G I +GN+S L +L
Sbjct: 565 AMPLLRHLHLGGNFFSGQIPPEYGTWQHLQYLALSGNELAGTIAPELGNLSSLRELYIG- 741
Query: 246 NTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTI 305
Y + SG IP + NL NL L LSG IP+ +
Sbjct: 742 -----------------------YYNTYSGGIPPEIGNLSNLVRLDAAYCGLSGEIPAEL 852
Query: 306 GNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVA 365
G L+NL L L+ N LSGS+ +G+L +LK + N L+G +PA+ LK L + +
Sbjct: 853 GKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGEVPASFAELKNLTLLNLF 1032
Query: 366 SNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSL 425
NKL+G IP +FVG LP+ L+ L + N FTG +P +L
Sbjct: 1033 RNKLHGAIP-----------------EFVGELPA-------LEVLQLWENNFTGSIPQNL 1140
Query: 426 KSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMIS 485
+ + + + N+I G + + L+ + N G I + GK L +
Sbjct: 1141 GNNGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSLGKCKSLNRIRMG 1320
Query: 486 NTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLL-YLKISNNHFTDSIPT 544
++G IP GL KL ++ L N LTG+ P++ G + + L + +SNN + S+P+
Sbjct: 1321 ENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPED--GSIATDLGQISLSNNQLSGSLPS 1494
Query: 545 EIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSA--LASID 602
IG +++L L GNE +G IP ++ L +L ++ S N+ G I L ID
Sbjct: 1495 TIGNFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDFSHNKFSGPIAPEISKCKLLTFID 1674
Query: 603 LSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFS--MSLDFVNISDNQLDGPLPE 660
LSGN L+G IP + + L+ LNLS N L G+IP + SL V+ S N G +P
Sbjct: 1675 LSGNELSGEIPNKITSMRILNYLNLSRNHLDGSIPGNIASMQSLTSVDFSYNNFSGLVPG 1854
Query: 661 NPAFLRAPFESFKNNKGLCGNITGLVPCATSQIHSRKSKNILQSVFIALGALILVLSGVG 720
F + SF N LCG G PC + + ++ + F + L+LV+ +
Sbjct: 1855 TGQFGYFNYTSFLGNPELCGPYLG--PCKDGVANGPRQPHV-KGPFSSSLKLLLVIGLLV 2025
Query: 721 ISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQ 780
S+ ++ E + L + D +++++ + + +IG G
Sbjct: 2026 CSILFAVAAIFKARALKKASEARAWKLTAFQRLD--FTVDDVLDCLK---EDNIIGKGGA 2190
Query: 781 GNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKS--FTSEIETLTGIKHRNIIKLHGFCS 838
G VYK +P G VAVK+L MS SS F +EI+TL I+HR+I++L GFCS
Sbjct: 2191 GIVYKGAMPNGDNVAVKRL-----PAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCS 2355
Query: 839 HSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHR 898
+ + + LVY++M GSL ++L+ +K W+ R + A L YLHHDCSP I+HR
Sbjct: 2356 NHETNLLVYEYMPNGSLGEVLHGKKGG-HLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHR 2532
Query: 899 DISSKNILLNLDYEAHVSDFGTAKFLKPDLHS--WTQFAGTFGYAAPELSQTMEVNEKCD 956
D+ S NILL+ ++EAHV+DFG AKFL+ S + AG++GY APE + T++V+EK D
Sbjct: 2533 DVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYTLKVDEKSD 2712
Query: 957 VYSFGVLALEIIIGKHP----GDLISLFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEE 1012
VYSFGV+ LE++ G+ P GD + + +N + +VLD R V P+ E
Sbjct: 2713 VYSFGVVLLELVTGRKPVGEFGDGVDIVQWVRKMTDSNKEGVLKVLDSRLPSV--PL-HE 2883
Query: 1013 VILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
V+ + +A C+ + RPTM +V ++L
Sbjct: 2884 VMHVFYVAMLCVEEQAVERPTMREVVQIL 2970
>TC229875 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat
receptor-like protein kinase 1, partial (23%)
Length = 1141
Score = 384 bits (985), Expect = e-106
Identities = 197/293 (67%), Positives = 232/293 (78%), Gaps = 4/293 (1%)
Frame = +2
Query: 753 HDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSS 812
H GKM+FENIIEATE+FDDK+LIGVG QG VYKA LPTG VVAVKKLH V + EM +
Sbjct: 2 HQGKMVFENIIEATEDFDDKHLIGVGGQGCVYKAVLPTGQVVAVKKLHSVPNGEM--LNL 175
Query: 813 KSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEK 872
K+FT EI+ LT I+HRNI+KL+GFCSHS+FSFLV +F+E GS+ + L ++ QA+AFDW K
Sbjct: 176 KAFTCEIQALTEIRHRNIVKLYGFCSHSQFSFLVCEFLENGSVGKTLKDDGQAMAFDWYK 355
Query: 873 RVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHSWT 932
RVNVVK VANAL Y+HH+CSP I+HRDISSKN+LL+ +Y AHVSDFGTAKFL PD +WT
Sbjct: 356 RVNVVKDVANALCYMHHECSPRIVHRDISSKNVLLDSEYVAHVSDFGTAKFLNPDSSNWT 535
Query: 933 QFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFL--SPST--RPT 988
F GTFGYAAPEL+ TMEVNEKCDVYSFGVLA EI+IGKHPGD+IS L SPST T
Sbjct: 536 SFVGTFGYAAPELAYTMEVNEKCDVYSFGVLAWEILIGKHPGDVISCLLGSSPSTLVAST 715
Query: 989 ANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
+ M L + LDQR KPI +EV IAK+A +CL + PRSRPTM+QV L
Sbjct: 716 LDHMALMDKLDQRLPHPTKPIGKEVASIAKIAMACLTESPRSRPTMEQVANEL 874
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 284 bits (726), Expect = 2e-76
Identities = 227/783 (28%), Positives = 357/783 (44%), Gaps = 30/783 (3%)
Frame = +2
Query: 289 VLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGT 348
V+ L L G I IG L+ L L L +N++ GSIP+++G L NL+ + N LTG+
Sbjct: 449 VIQLPWKGLRGRITDKIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGS 628
Query: 349 IPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLK 408
IP ++G L ++++N L G IP L N T Y +S N F G LP+ + SL
Sbjct: 629 IPLSLGFCPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLT 808
Query: 409 YLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGH 468
+LS +N +G +P S GN G + L+ + L N F G
Sbjct: 809 FLSLQNNNLSGSLPNSW-----------GGNSKNG--------FFRLQNLILDHNFFTGD 931
Query: 469 ISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSL 528
+ + G +L +S+ SG IP + L++L L +S+N L G LP L + SL
Sbjct: 932 VPASLGSLRELNEISLSHNKFSGAIPNEIGTLSRLKTLDISNNALNGNLP-ATLSNLSSL 1108
Query: 529 LYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEG 588
L NN + IP +G L+ L L L N+ SG IP+ +A + LR L+LS N G
Sbjct: 1109 TLLNAENNLLDNQIPQSLGRLRNLSVLILSRNQFSGHIPSSIANISSLRQLDLSLNNFSG 1288
Query: 589 RIPSTFDSALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMSLDFVN 648
IP +FDS + L++ N+S+N LSG++P + + +
Sbjct: 1289 EIPVSFDSQRS----------------------LNLFNVSYNSLSGSVPPLLAKKFNSSS 1402
Query: 649 ISDN-QLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCATSQIHSRKSKNILQSVFI 707
N QL G P P F +AP ++G+ P S+ H + + + +
Sbjct: 1403 FVGNIQLCGYSPSTPCFSQAP------SQGVIA-----PPPEVSKHHHHRKLSTKDIILV 1549
Query: 708 ALGALILVLSGV-GISMYVFFRRKKPNEEIQTEEEVQKGVLF--------SIW------- 751
G L++VL + + ++ R++ ++ + + V + S W
Sbjct: 1550 VAGVLLVVLIILCCVLLFCLIRKRSTSKAGNGQATEGRAVTYEDRKRSPSSCWWLMLKQV 1729
Query: 752 --------SHDGKMMF--ENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHL 801
DG M F ++++ AT ++G + G VYKA L G VAVK+L
Sbjct: 1730 GRLEGNLVHFDGPMAFTADDLLCATAE-----IMGKSTNGTVYKAILEDGSQVAVKRL-- 1888
Query: 802 VRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSK-FSFLVYKFMEGGSLDQILN 860
E + F SE+ L I+H N++ L + K LV+ +M GSL L+
Sbjct: 1889 ---REKIAKGHREFESEVSVLGKIRHPNVLALRAYYLGPKGEKLLVFDYMSKGSLASFLH 2059
Query: 861 NEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGT 920
DW R+ + + +A L LH IIH +++S N+LL+ + A ++DFG
Sbjct: 2060 GGGTETFIDWPTRMKIAQDLARGLFCLH--SQENIIHGNLTSSNVLLDENTNAKIADFGL 2233
Query: 921 AKFLKPDLHS-WTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPG-DLIS 978
++ + +S AG GY APELS+ + N K D+YS GV+ LE++ K PG +
Sbjct: 2234 SRLMSTTANSNVIATAGALGYRAPELSKLKKANTKTDIYSLGVILLELLTRKSPGVSMNG 2413
Query: 979 LFLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVC 1038
L L + EV D + + +E++ KLA C++ P +RP + QV
Sbjct: 2414 LDLPQWVASVVKEEWTNEVFDADLMRDASTVGDELLNTLKLALHCVDPSPSARPEVHQVL 2593
Query: 1039 KML 1041
+ L
Sbjct: 2594 QQL 2602
Score = 180 bits (456), Expect = 3e-45
Identities = 109/300 (36%), Positives = 155/300 (51%), Gaps = 5/300 (1%)
Frame = +2
Query: 151 LSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGL 210
L G I IG L L L L N GG IP +G L LR + + L GSIP +G
Sbjct: 473 LRGRITDKIGQLQGLRKLSLHDNQI-GGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGF 649
Query: 211 LTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYN 270
L +DLSNN L+G IP ++ N +KL L + N+ GP+P SL + SLT + L N
Sbjct: 650 CPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNS-FSGPLPASLTHSFSLTFLSLQN 826
Query: 271 MSLSGSIPDS-----VQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSI 325
+LSGS+P+S L L L N +G +P+++G+L+ L + L +N+ SG+I
Sbjct: 827 NNLSGSLPNSWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAI 1006
Query: 326 PASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYS 385
P IG L LK + N L G +PAT+ NL L + +N L +IP L + N
Sbjct: 1007PNEIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSV 1186
Query: 386 FVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDI 445
++S N F GH+PS + SL+ L N F+G +P S S S+ + N + G +
Sbjct: 1187LILSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVSFDSQRSLNLFNVSYNSLSGSV 1366
Score = 153 bits (387), Expect = 3e-37
Identities = 123/407 (30%), Positives = 186/407 (45%), Gaps = 13/407 (3%)
Frame = +2
Query: 5 TFIIMILCVLPTL-------SVAEDSEAKLALLKWKASFDNQSQSILSTWKNTT-NPCSK 56
+ +++ CV P L V + LAL +K + + L +W ++ CS
Sbjct: 233 SLVVLPSCVRPVLCEDEGWDGVVVTASNLLALEAFKQELADP-EGFLRSWNDSGYGACSG 409
Query: 57 -WRGIECDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLS 115
W GI+C + +I I L GL+G I +IG L
Sbjct: 410 GWVGIKCAQGQVI-VIQLPWKGLRGR-------------------------ITDKIGQLQ 511
Query: 116 RINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNF 175
+ L+ N I GSIP + L +L+G+ F L+G I S+G L LDL NN
Sbjct: 512 GLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFCPLLQSLDLS-NNL 688
Query: 176 SGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNM 235
G IP + KL +L ++ S G +P + +LT++ L NN LSG +P + G
Sbjct: 689 LTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLTFLSLQNNNLSGSLPNSWGGN 868
Query: 236 SKLN----QLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLA 291
SK Q + ++ G +P SL ++ L I L + SG+IP+ + L L L
Sbjct: 869 SKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAIPNEIGTLSRLKTLD 1048
Query: 292 LYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPA 351
+ N L+G +P+T+ NL +LTLL NN L IP S+G L NL + N +G IP+
Sbjct: 1049ISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLILSRNQFSGHIPS 1228
Query: 352 TIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLP 398
+I N+ L +++ N G IP + + F VS N G +P
Sbjct: 1229SIANISSLRQLDLSLNNFSGEIPVSFDSQRSLNLFNVSYNSLSGSVP 1369
>TC234472 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat
receptor-like protein kinase 1, partial (16%)
Length = 553
Score = 262 bits (670), Expect = 5e-70
Identities = 121/178 (67%), Positives = 159/178 (88%)
Frame = +1
Query: 747 LFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEE 806
LF+IWS DGK+++ENI+EATE+FD+K+LIGVG QG+VYKA+L TG ++AVKKLHLV++ E
Sbjct: 22 LFAIWSFDGKLVYENIVEATEDFDNKHLIGVGGQGSVYKAKLHTGQILAVKKLHLVQNGE 201
Query: 807 MSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAI 866
+S + K+FTSEI+ L I+HRNI+KL+GFCSHS+ SFLVY+F+E GS+D+IL +++QAI
Sbjct: 202 LS--NIKAFTSEIQALINIRHRNIVKLYGFCSHSQSSFLVYEFLEKGSIDKILKDDEQAI 375
Query: 867 AFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFL 924
AFDW+ R+N +KGVANALSY+HHDCSPPI+HRDISSKNI+L+L+Y AHVSDFG A+ L
Sbjct: 376 AFDWDPRINAIKGVANALSYMHHDCSPPIVHRDISSKNIVLDLEYVAHVSDFGAARLL 549
>TC209281 similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat receptor-like
protein kinase 1, partial (14%)
Length = 586
Score = 238 bits (606), Expect = 1e-62
Identities = 119/193 (61%), Positives = 149/193 (76%), Gaps = 3/193 (1%)
Frame = +2
Query: 848 KFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILL 907
+F+E G + +IL +++QAIA DW KRV++VKGVANAL Y+HHDCSPPI+HRDISSKN+LL
Sbjct: 8 EFLEKGDVKKILKDDEQAIALDWNKRVDIVKGVANALCYMHHDCSPPIVHRDISSKNVLL 187
Query: 908 NLDYEAHVSDFGTAKFLKPDLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEI 967
+ D AHV+DFGTAKFL PD +WT FAGT+GYAAPEL+ TME NEKCDVYSFGV ALEI
Sbjct: 188 DSDDVAHVADFGTAKFLNPDSSNWTSFAGTYGYAAPELAYTMEANEKCDVYSFGVFALEI 367
Query: 968 IIGKHPGDLI-SLFLSPSTRPTA--NDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCL 1024
+ G+HPGD+ SL LS S+ T+ + M L LD+R PID+EVI I K+A + L
Sbjct: 368 LFGEHPGDVTSSLLLSSSSTMTSTLDHMSLMVKLDERLPHPTSPIDKEVISIVKIAIARL 547
Query: 1025 NQVPRSRPTMDQV 1037
+ RSRP+M+QV
Sbjct: 548 TESTRSRPSMEQV 586
>TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase PERK1-like,
partial (80%)
Length = 1604
Score = 226 bits (575), Expect = 5e-59
Identities = 141/407 (34%), Positives = 214/407 (51%), Gaps = 5/407 (1%)
Frame = +1
Query: 642 MSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCGNITGLVPCATSQIHSRKSKNI 701
+SL +N+S N+L G +P + F R P +SF N GLCGN L PC ++ R + +
Sbjct: 1 LSLSLLNVSYNKLFGVIPTSNNFTRFPPDSFIGNPGLCGNWLNL-PCHGARPSERVTLSK 177
Query: 702 LQSVFIALGALILVLSGVGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFEN 761
+ I LGAL+++L + + P+ L + + ++E+
Sbjct: 178 AAILGITLGALVILLMVLVAACRPHSPSPFPDGSFDKPINFSPPKLVILHMNMALHVYED 357
Query: 762 IIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIET 821
I+ TEN +KY+IG G+ VYK L VA+K+++ + + K F +E+ET
Sbjct: 358 IMRMTENLSEKYIIGYGASSTVYKCVLKNCKPVAIKRIYSHYPQCI-----KEFETELET 522
Query: 822 LTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVA 881
+ IKHRN++ L G+ L Y +ME GSL +L+ + DWE R+ + G A
Sbjct: 523 VGSIKHRNLVSLQGYSLSPYGHLLFYDYMENGSLWDLLHGPTKKKKLDWELRLKIALGAA 702
Query: 882 NALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD-LHSWTQFAGTFGY 940
L+YLHHDC P IIHRD+ S NILL+ D+E H++DFG AK L P H+ T GT GY
Sbjct: 703 QGLAYLHHDCCPRIIHRDVKSSNILLDADFEPHLTDFGIAKSLCPSKSHTSTYIMGTIGY 882
Query: 941 AAPELSQTMEVNEKCDVYSFGVLALEIIIGK----HPGDLISLFLSPSTRPTANDMLLTE 996
PE ++T + EK DVYS+G++ LE++ G+ + +L L LS A + E
Sbjct: 883 IDPEYARTSHLTEKSDVYSYGIVLLELLTGRKAVDNESNLHHLILS-----KAATNAVME 1047
Query: 997 VLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKMLGA 1043
+D K + V + +LA C + P RPTM +V ++LG+
Sbjct: 1048 TVDPDITATCKDLG-AVKKVYQLALLCTKRQPADRPTMHEVTRVLGS 1185
>TC231717 weakly similar to UP|Q8VZG8 (Q8VZG8) AT4g08850/T32A17_160, partial
(11%)
Length = 453
Score = 199 bits (506), Expect = 5e-51
Identities = 92/150 (61%), Positives = 121/150 (80%)
Frame = +3
Query: 783 VYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKF 842
VYKA+LP G +VAVKKLH +EE SK+F++E++ L IKHRNI+K G+C H +F
Sbjct: 6 VYKAKLPAGQIVAVKKLHAAPNEETP--DSKAFSTEVKALAEIKHRNIVKSLGYCLHPRF 179
Query: 843 SFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISS 902
SFL+Y+F+EGGSLD++L ++ +A FDWE+RV VVKGVA+AL ++HH C PPI+HRDISS
Sbjct: 180 SFLIYEFLEGGSLDKVLTDDTRATMFDWERRVKVVKGVASALYHMHHGCFPPIVHRDISS 359
Query: 903 KNILLNLDYEAHVSDFGTAKFLKPDLHSWT 932
KN+L++LDYEAH+SDFGTAK LKPD + T
Sbjct: 360 KNVLIDLDYEAHISDFGTAKILKPDSQNIT 449
>TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-like, partial
(31%)
Length = 1329
Score = 199 bits (505), Expect = 7e-51
Identities = 157/503 (31%), Positives = 229/503 (45%), Gaps = 3/503 (0%)
Frame = +1
Query: 302 PSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIV 361
PS + N++ LT L L +N +GSIP S +L NL+ SV N+++GT+P I L L
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 362 FEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPV 421
+ +NK G +P L + S ND VG++P +C G L L F N+FTG +
Sbjct: 181 LLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGGL 360
Query: 422 PTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLET 481
+S+ +CSS+ R+R+E N G+I F + P++ YVDLS N F G I + ++ LE
Sbjct: 361 -SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEY 537
Query: 482 FMIS-NTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTD 540
F +S N + G IP L +L SS ++ LP + KS+
Sbjct: 538 FNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLP--LFESCKSI------------ 675
Query: 541 SIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDS--AL 598
+DL N LSGTIPN V++ L +NLS N + G IP S L
Sbjct: 676 ------------SVIDLDSNSLSGTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVL 819
Query: 599 ASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMSLDFVNISDNQLDGPL 658
+DLS N+ NG IP G L +LN+S N +SG+IP+ S L
Sbjct: 820 GVVDLSNNKFNGPIPAKFGSSSNLQLLNVSFNNISGSIPTGKSFKL-------------- 957
Query: 659 PENPAFLRAPFESFKNNKGLCGNITGLVPCATSQIHSRKSKNILQSVFIALGALILVLSG 718
+F N LCG P + + S+ S + + V +++G L++VL G
Sbjct: 958 --------MGRSAFVGNSELCGAPLQPCPDSVGILGSKCSWKVTRIVLLSVG-LLIVLLG 1110
Query: 719 VGISMYVFFRRKKPNEEIQTEEEVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVG 778
+ M R K ++ V L ++D + TE
Sbjct: 1111LAFGMSYLRRGIKSQWKM-----VSFAGLPQFTANDVLTSLSATTKPTE----------V 1245
Query: 779 SQGNVYKAELPTGLVVAVKKLHL 801
+V KA LPTG+ V VKK+ L
Sbjct: 1246QSPSVTKAVLPTGITVLVKKIEL 1314
Score = 138 bits (347), Expect = 1e-32
Identities = 105/341 (30%), Positives = 159/341 (45%), Gaps = 30/341 (8%)
Frame = +1
Query: 254 PHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTL 313
P L N+ LT + L + +GSIP+S +L NL +L++ N++SG +P I L +L
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 314 LLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATI---GNLKQLIVFEVASNKLY 370
LL+ NN+ SGS+P S+G LK+ N+L G IP I G L +LI+F SNK
Sbjct: 181 LLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILF---SNKFT 351
Query: 371 GRIPNGLYNITNWYSFV---VSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKS 427
G GL +I+N S V + +N F G + + + Y+ N F G +P+ +
Sbjct: 352 G----GLSSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQ 519
Query: 428 CSSIERIRIEGNQIEGDI---------------AEDFGVYPNL---------RYVDLSDN 463
+ +E + NQ G I A G+ +L +DL N
Sbjct: 520 ATQLEYFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPLFESCKSISVIDLDSN 699
Query: 464 KFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILG 523
G I K LE +SN N++G IP + + LG + LS+N+ G +P + G
Sbjct: 700 SLSGTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAK-FG 876
Query: 524 GMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSG 564
+L L +S N+ + SIPT +G +EL G
Sbjct: 877 SSSNLQLLNVSFNNISGSIPTGKSFKLMGRSAFVGNSELCG 999
Score = 124 bits (310), Expect = 3e-28
Identities = 90/309 (29%), Positives = 140/309 (45%)
Frame = +1
Query: 68 ISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPI 127
++ +DL++ G++ +FS NL L++ N GT+P I L + TL N
Sbjct: 28 LTDLDLSDNFFTGSIPE-SFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLETLLIWNNKF 204
Query: 128 IGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKL 187
GS+P+ + LK +D L G I I L L L N F+GG I
Sbjct: 205 SGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGGL--SSISNC 378
Query: 188 KKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNT 247
L L + G I + LL ++ Y+DLS N G IP I ++L + N
Sbjct: 379 SSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEYFNVSYNQ 558
Query: 248 KLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGN 307
+L G IP W++ L + +S +P ++ ++ V+ L N+LSG IP+ +
Sbjct: 559 QLGGIIPSQTWSLPQLQNFSASSCGISSDLP-LFESCKSISVIDLDSNSLSGTIPNGVSK 735
Query: 308 LKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASN 367
+ L + L NN L+G IP + ++ L + N G IPA G+ L + V+ N
Sbjct: 736 CQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSNLQLLNVSFN 915
Query: 368 KLYGRIPNG 376
+ G IP G
Sbjct: 916 NISGSIPTG 942
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 197 bits (500), Expect = 3e-50
Identities = 115/295 (38%), Positives = 174/295 (58%), Gaps = 10/295 (3%)
Frame = +1
Query: 756 KMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSF 815
K+ F ++++AT F + LIG G G+VYKA+L G VVA+KKL V + + F
Sbjct: 43 KLTFADLLDATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQ-----GDREF 207
Query: 816 TSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQA-IAFDWEKRV 874
T+E+ET+ IKHRN++ L G+C + LVY++M+ GSL+ +L+++K+A I +W R
Sbjct: 208 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQKKAGIKLNWAIRR 387
Query: 875 NVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKP-DLH-SWT 932
+ G A L++LHH+C P IIHRD+ S N+LL+ + EA VSDFG A+ + D H S +
Sbjct: 388 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 567
Query: 933 QFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISL-------FLSPST 985
AGT GY PE Q+ + K DVYS+GV+ LE++ GK P D ++
Sbjct: 568 TLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSADFGDNNLVGWVKQHA 747
Query: 986 RPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKM 1040
+ +D+ E++ + P ++ E++ K+A SCL+ P RPTM QV M
Sbjct: 748 KLKISDIFDPELMKEDPN-----LEMELLQHLKIAVSCLDDRPWRRPTMIQVMAM 897
>TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial
(14%)
Length = 1790
Score = 192 bits (489), Expect = 5e-49
Identities = 176/596 (29%), Positives = 279/596 (46%), Gaps = 36/596 (6%)
Frame = +1
Query: 98 IYNNHFY-GTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEID 156
IY FY G IP I +L I L+ N + G +P + L+ L+ LD T + I
Sbjct: 1 IYTVTFYRGKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIP 180
Query: 157 KSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTY 216
NL++L L+L N +G IP LK L+ L + SL G +P +G L+NL
Sbjct: 181 SPFANLSSLRTLNLAHNRLNG-TIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVT 357
Query: 217 IDLSNNFLSGVIPETIGNMSKLNQL--MFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLS 274
+DLS+N L G I E+ N KL L + + T L+ + L + L + +
Sbjct: 358 LDLSSNLLEGSIKES--NFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIG 531
Query: 275 GSIPDSVQNLINLDVLALYMNNLSGFIPST--IGNLKNLTLLLLRNNRLSGSIPASIGNL 332
P+ ++ ++ VL + ++ +PS I L+ + L L NN L G +
Sbjct: 532 PKFPEWLKRQSSVKVLTMSKAGIADLVPSWFWIWTLQ-IEFLDLSNNLLRGDLS---NIF 699
Query: 333 INLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPN---GLYNITNWYSFV-V 388
+N ++ N G +P+ N++ V VA+N + G I G N TN S +
Sbjct: 700 LNSSVINLSSNLFKGRLPSVSANVE---VLNVANNSISGTISPFLCGKPNATNKLSVLDF 870
Query: 389 SENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAED 448
S N G L +L +++ N +G +P S+ S +E + ++ N+ G I
Sbjct: 871 SNNVLSGDLGHCWVHWQALVHVNLGSNNMSGEIPNSMGYLSQLESLLLDDNRFSGYIPST 1050
Query: 449 FGVYPNLRYVDLSDNKFHGHISPNWGKSLD-LETFMISNTNISGGIPLDFIGLTKLGRLH 507
++++D+ +N+ I P+W + L + + N +G I L+ L L
Sbjct: 1051LQNCSTMKFIDMGNNQLSDTI-PDWMWEMQYLMVLRLRSNNFNGSITQKMCQLSSLIVLD 1227
Query: 508 LSSNQLTGKLPKEILGGMKSLLY--------------LKISNNHFTDS---IPTEIGLLQ 550
+N L+G +P L MK++ S NH+ ++ +P + L
Sbjct: 1228HGNNSLSGSIPN-CLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLVPKKDELEY 1404
Query: 551 R-----LEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSA--LASIDL 603
R + +DL N+LSG IP+E+++L LR LNLSRN + G IP+ L S+DL
Sbjct: 1405RDNLILVRMIDLSSNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKMKLLESLDL 1584
Query: 604 SGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIP-STFSMSLDFVNISDN-QLDGP 657
S N ++G IP SL L LS LNLS++ LSG IP ST S + ++ + N +L GP
Sbjct: 1585SLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRIPTSTQLQSFEELSYTGNPELCGP 1752
Score = 185 bits (470), Expect = 8e-47
Identities = 167/583 (28%), Positives = 259/583 (43%), Gaps = 104/583 (17%)
Frame = +1
Query: 202 GSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMS 261
G IPQ I L N+ +DL NN LSG +P+++G + L L +NNT PIP N+S
Sbjct: 25 GKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNT-FTCPIPSPFANLS 201
Query: 262 SLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRL 321
SL + L + L+G+IP S + L NL VL L N+L+G +P T+G L NL L L +N L
Sbjct: 202 SLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLSSNLL 381
Query: 322 SGSIPAS-IGNLINLKYFSVQVNNL-----TGTIP------------------------- 350
GSI S L LK + NL +G P
Sbjct: 382 EGSIKESNFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKRQ 561
Query: 351 ----------ATIGNLK---------QLIVFEVASNKLYGRIPNGLYNITNWYSFVVSEN 391
A I +L Q+ ++++N L G + N N + +S N
Sbjct: 562 SSVKVLTMSKAGIADLVPSWFWIWTLQIEFLDLSNNLLRGDLSNIFLNSS---VINLSSN 732
Query: 392 DFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSL----KSCSSIERIRIEGNQIEGDIAE 447
F G LPS +++ L+ +N +G + L + + + + N + GD+
Sbjct: 733 LFKGRLPS---VSANVEVLNVANNSISGTISPFLCGKPNATNKLSVLDFSNNVLSGDLGH 903
Query: 448 DFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLH 507
+ + L +V+L N G I + G LE+ ++ + SG IP + + +
Sbjct: 904 CWVHWQALVHVNLGSNNMSGEIPNSMGYLSQLESLLLDDNRFSGYIPSTLQNCSTMKFID 1083
Query: 508 LSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIP 567
+ +NQL+ +P + + M+ L+ L++ +N+F SI ++ L L LD G N LSG+IP
Sbjct: 1084MGNNQLSDTIP-DWMWEMQYLMVLRLRSNNFNGSITQKMCQLSSLIVLDHGNNSLSGSIP 1260
Query: 568 NEVAEL----------------------------------PK------------LRMLNL 581
N + ++ PK +RM++L
Sbjct: 1261NCLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLVPKKDELEYRDNLILVRMIDL 1440
Query: 582 SRNRIEGRIPSTFD--SALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPST 639
S N++ G IPS SAL ++LS N L+G IP +G + L L+LS N +SG IP +
Sbjct: 1441SSNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQIPQS 1620
Query: 640 FS--MSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLCG 680
S L F+N+S + L G +P + S+ N LCG
Sbjct: 1621LSDLSFLSFLNLSYHNLSGRIPTSTQLQSFEELSYTGNPELCG 1749
Score = 152 bits (383), Expect = 1e-36
Identities = 151/579 (26%), Positives = 250/579 (43%), Gaps = 82/579 (14%)
Frame = +1
Query: 68 ISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPI 127
I +DL N L G L + +L L++ NN F IP NLS + TLN + N +
Sbjct: 61 IKNLDLQNNQLSGPLPD-SLGQLKHLEVLDLSNNTFTCPIPSPFANLSSLRTLNLAHNRL 237
Query: 128 IGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGN-------------- 173
G+IP+ L++L+ L+ +L+G++ ++G L+NL LDL N
Sbjct: 238 NGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLSSNLLEGSIKESNFVKL 417
Query: 174 ---------------------------------NFSGGPIPPE-IGKLKKLRYLAITQGS 199
+F GP PE + + ++ L +++
Sbjct: 418 FTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKRQSSVKVLTMSKAG 597
Query: 200 LVGSIPQEIGLLT-NLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLW 258
+ +P + T + ++DLSNN L G + N S +N ++ G +P
Sbjct: 598 IADLVPSWFWIWTLQIEFLDLSNNLLRGDLSNIFLNSSVIN----LSSNLFKGRLPSVSA 765
Query: 259 NMSSLTLIYLYNMSLSGSI-------PDSVQNLINLDV---------------------L 290
N+ L + N S+SG+I P++ L LD +
Sbjct: 766 NVEVLNVA---NNSISGTISPFLCGKPNATNKLSVLDFSNNVLSGDLGHCWVHWQALVHV 936
Query: 291 ALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIP 350
L NN+SG IP+++G L L LLL +NR SG IP+++ N +K+ + N L+ TIP
Sbjct: 937 NLGSNNMSGEIPNSMGYLSQLESLLLDDNRFSGYIPSTLQNCSTMKFIDMGNNQLSDTIP 1116
Query: 351 ATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYL 410
+ ++ L+V + SN G I + +++ N G +P+ + +K +
Sbjct: 1117DWMWEMQYLMVLRLRSNNFNGSITQKMCQLSSLIVLDHGNNSLSGSIPNCL---DDMKTM 1287
Query: 411 SAFHNRFTGPVPTSLKSCSSI----ERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFH 466
+ + F P S S S E + + + E + ++ + +R +DLS NK
Sbjct: 1288AGEDDFFANPSSYSYGSDFSYNHYKETLVLVPKKDELEYRDNLIL---VRMIDLSSNKLS 1458
Query: 467 GHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMK 526
G I K L +S ++SG IP D + L L LS N ++G++P+ L +
Sbjct: 1459GAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQIPQS-LSDLS 1635
Query: 527 SLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGN-ELSG 564
L +L +S ++ + IPT LQ EEL GN EL G
Sbjct: 1636FLSFLNLSYHNLSGRIPTST-QLQSFEELSYTGNPELCG 1749
Score = 89.0 bits (219), Expect = 1e-17
Identities = 74/270 (27%), Positives = 128/270 (47%), Gaps = 28/270 (10%)
Frame = +1
Query: 417 FTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKS 476
+ G +P + S +I+ + ++ NQ+ G + + G +L +DLS+N F I +
Sbjct: 19 YRGKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIPSPFANL 198
Query: 477 LDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNN 536
L T +++ ++G IP F L L L+L +N LTG +P LG + +L+ L +S+N
Sbjct: 199 SSLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVP-VTLGTLSNLVTLDLSSN 375
Query: 537 HFTDSI-PTEIGLLQRLEELDLGGNEL-----SGTIPNEVAEL---------PK------ 575
SI + L L+EL L L SG P E PK
Sbjct: 376 LLEGSIKESNFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLK 555
Query: 576 ----LRMLNLSRNRIEGRIPSTF---DSALASIDLSGNRLNGNIPTSLGFLVQLSMLNLS 628
+++L +S+ I +PS F + +DLS N L G++ + S++NLS
Sbjct: 556 RQSSVKVLTMSKAGIADLVPSWFWIWTLQIEFLDLSNNLLRGDLS---NIFLNSSVINLS 726
Query: 629 HNMLSGTIPSTFSMSLDFVNISDNQLDGPL 658
N+ G +PS S +++ +N+++N + G +
Sbjct: 727 SNLFKGRLPSV-SANVEVLNVANNSISGTI 813
>TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial (5%)
Length = 1279
Score = 189 bits (481), Expect = 4e-48
Identities = 103/233 (44%), Positives = 143/233 (61%), Gaps = 6/233 (2%)
Frame = +1
Query: 815 FTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRV 874
F E+E L IKHR ++ L G+C+ L+Y ++ GGSLD+ L+ ++A DW+ R+
Sbjct: 46 FERELEILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEALH--ERADQLDWDSRL 219
Query: 875 NVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLK-PDLHSWTQ 933
N++ G A L+YLHHDCSP IIHRDI S NILL+ + EA VSDFG AK L+ + H T
Sbjct: 220 NIIMGAAKGLAYLHHDCSPRIIHRDIKSSNILLDGNLEARVSDFGLAKLLEDEESHITTI 399
Query: 934 FAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRPTANDML 993
AGTFGY APE Q+ EK DVYSFGVL LE++ GK P D + P+ N L
Sbjct: 400 VAGTFGYLAPEYMQSGRATEKSDVYSFGVLTLEVLSGKRPTDAAFIEKGPNIVGWLN-FL 576
Query: 994 LTEVLDQRPQKVIKPIDEEVIL-----IAKLAFSCLNQVPRSRPTMDQVCKML 1041
+TE RP++++ P+ E V + + +A C++ P RPTM +V ++L
Sbjct: 577 ITE---NRPREIVDPLCEGVQMESLDALLSVAIQCVSSSPEDRPTMHRVVQLL 726
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 174 bits (440), Expect = 2e-43
Identities = 115/329 (34%), Positives = 168/329 (50%), Gaps = 13/329 (3%)
Frame = +2
Query: 726 FFRRKKPNE---EIQTEE--EVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQ 780
++RR+KP + ++ EE EV G L + + AT+NF +K+++G G
Sbjct: 32 YWRRRKPQDHFFDVPAEEDPEVHLGQL-------KRFSLRELQVATDNFSNKHILGRGGF 190
Query: 781 GNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHS 840
G VYK L G +VAVK+L EE + F +E+E ++ HRN+++L GFC
Sbjct: 191 GKVYKGRLADGSLVAVKRLK----EERTQGG*LQFQTEVEMISMAVHRNLLRLRGFCMTP 358
Query: 841 KFSFLVYKFMEGGSLDQILNNEKQAIA-FDWEKRVNVVKGVANALSYLHHDCSPPIIHRD 899
LVY +M GS+ L +++ W +R + G A L+YLH C P IIHRD
Sbjct: 359 TERLLVYPYMANGSVASCLRERQESQPPLGWPERKRIALGSARGLAYLHDHCDPKIIHRD 538
Query: 900 ISSKNILLNLDYEAHVSDFGTAKFLK-PDLHSWTQFAGTFGYAAPELSQTMEVNEKCDVY 958
+ + NILL+ ++EA V DFG AK + D H T GT G+ APE T + +EK DV+
Sbjct: 539 VKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVF 718
Query: 959 SFGVLALEIIIGKHPGDLISL------FLSPSTRPTANDMLLTEVLDQRPQKVIKPIDEE 1012
+GV+ LE+I G+ DL L L + D L ++D Q DEE
Sbjct: 719 GYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKDRKLETLVDADLQGSYN--DEE 892
Query: 1013 VILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
V + ++A C P RP M +V +ML
Sbjct: 893 VEQLIQVALLCTQGSPMERPKMSEVVRML 979
>CO979898
Length = 869
Score = 174 bits (440), Expect = 2e-43
Identities = 91/210 (43%), Positives = 122/210 (57%), Gaps = 1/210 (0%)
Frame = -1
Query: 767 ENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIK 826
+ + K +IG G G VY+ +L +A+K+L+ E K F E+E + IK
Sbjct: 869 QKLNSKDIIGSGGYGVVYELKLDDSTALAIKRLNRGTAER-----DKGFERELEAMADIK 705
Query: 827 HRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSY 886
HRNI+ LHG+ + ++ L+Y+ M GSLD L+ + DW R + G A +SY
Sbjct: 704 HRNIVTLHGYYTAPLYNLLIYELMPHGSLDSFLHGRSREKVLDWPTRYRIAAGAARGISY 525
Query: 887 LHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD-LHSWTQFAGTFGYAAPEL 945
LHHDC P IIHRDI S NILL+ + +A VSDFG A ++P+ H T AGTFGY APE
Sbjct: 524 LHHDCIPHIIHRDIKSSNILLDQNMDARVSDFGLATLMQPNKTHVSTIVAGTFGYLAPEY 345
Query: 946 SQTMEVNEKCDVYSFGVLALEIIIGKHPGD 975
T K DVYSFGV+ LE++ GK P D
Sbjct: 344 FDTGRATLKGDVYSFGVVLLELLTGKKPSD 255
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor kinase
1, partial (60%)
Length = 1444
Score = 171 bits (434), Expect = 1e-42
Identities = 119/338 (35%), Positives = 171/338 (50%), Gaps = 20/338 (5%)
Frame = +1
Query: 724 YVFFRRKKPNE---EIQTEE--EVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVG 778
+ ++RR+KP E ++ EE EV G L + + AT++F +K ++G G
Sbjct: 28 FAWWRRRKPQEFFFDVPAEEDPEVHLGQL-------KRFSLRELQVATDSFSNKNILGRG 186
Query: 779 SQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCS 838
G VYK L G +VAVK+L EE + F +E+E ++ HRN+++L GFC
Sbjct: 187 GFGKVYKGRLADGSLVAVKRLK----EERTPGGELQFQTEVEMISMAVHRNLLRLRGFCM 354
Query: 839 HSKFSFLVYKFMEGGSLDQILNNEKQ-AIAFDWEKRVNVVKGVANALSYLHHDCSPPIIH 897
LVY +M GS+ L DW R V G A LSYLH C P IIH
Sbjct: 355 TPTERLLVYPYMANGSVASCLRERPPYQEPLDWPTRKRVALGSARGLSYLHDHCDPKIIH 534
Query: 898 RDISSKNILLNLDYEAHVSDFGTAKFLK-PDLHSWTQFAGTFGYAAPELSQTMEVNEKCD 956
RD+ + NILL+ ++EA V DFG AK + D H T GT G+ APE T + +EK D
Sbjct: 535 RDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTD 714
Query: 957 VYSFGVLALEIIIGKHPGDLISLFLSPSTRPTAND---MLLTEV----LDQRPQKVIKP- 1008
V+ +G++ LE+I G+ DL L AND MLL V +++ + ++ P
Sbjct: 715 VFGYGIMLLELITGQRAFDLARL---------ANDDDVMLLDWVKGLLKEKKLEMLVDPD 867
Query: 1009 -----IDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
I+ EV + ++A C P RP M +V +ML
Sbjct: 868 LQTNYIETEVEQLIQVALLCTQGSPMDRPKMSEVVRML 981
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein ,
partial (47%)
Length = 1305
Score = 170 bits (430), Expect = 3e-42
Identities = 102/289 (35%), Positives = 151/289 (51%), Gaps = 6/289 (2%)
Frame = +2
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSE 818
F I AT+ F + +G G G VYK LP+G VAVK+L + + + F +E
Sbjct: 50 FSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQ-----GGEEFKNE 214
Query: 819 IETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVK 878
+E + ++HRN+++L GFC + LVY+F+ SLD IL + ++ + DW +R +V+
Sbjct: 215 VEIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVE 394
Query: 879 GVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD--LHSWTQFAG 936
G+A + YLH D IIHRD+ + N+LL+ D +SDFG A+ D + + G
Sbjct: 395 GIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVG 574
Query: 937 TFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRPTA----NDM 992
T+GY +PE + E + K DVYSFGVL LEII GK ++ A D
Sbjct: 575 TYGYMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLWKDE 754
Query: 993 LLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
E++DQ ++ EVI + C+ + P RPTM V ML
Sbjct: 755 APLELMDQSLRE--SYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLML 895
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 169 bits (429), Expect = 5e-42
Identities = 103/289 (35%), Positives = 152/289 (51%), Gaps = 6/289 (2%)
Frame = +3
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSE 818
F + AT F + G G+V++ LP G V+AVK+ L + K F SE
Sbjct: 12 FSELQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLASTQ-----GDKEFCSE 176
Query: 819 IETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVK 878
+E L+ +HRN++ L GFC LVY+++ GSLD + KQ + +W R +
Sbjct: 177 VEVLSCAQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRKQNV-LEWSARQKIAV 353
Query: 879 GVANALSYLHHDCSPP-IIHRDISSKNILLNLDYEAHVSDFGTAKFLKP-DLHSWTQFAG 936
G A L YLH +C I+HRD+ NILL D+EA V DFG A++ D+ T+ G
Sbjct: 354 GAARGLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQPDGDMGVETRVIG 533
Query: 937 TFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLI----SLFLSPSTRPTANDM 992
TFGY APE +Q+ ++ EK DVYSFG++ LE++ G+ D+ LS RP
Sbjct: 534 TFGYLAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEWARPLLEKQ 713
Query: 993 LLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
+++D + +D+EV + K + C+ + P RP M QV +ML
Sbjct: 714 ATYKLIDPSLRNCY--VDQEVYRMLKCSSLCIGRDPHLRPRMSQVLRML 854
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,444,943
Number of Sequences: 63676
Number of extensions: 657493
Number of successful extensions: 7790
Number of sequences better than 10.0: 1100
Number of HSP's better than 10.0 without gapping: 4390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5779
length of query: 1061
length of database: 12,639,632
effective HSP length: 107
effective length of query: 954
effective length of database: 5,826,300
effective search space: 5558290200
effective search space used: 5558290200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC133779.6


