

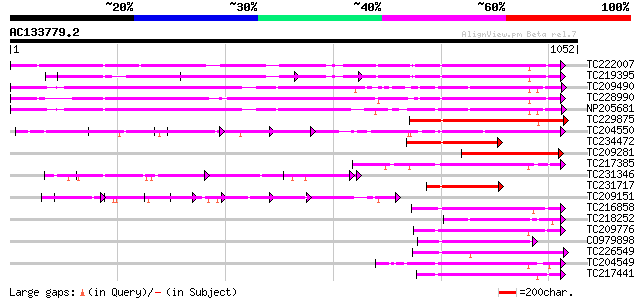
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.2 - phase: 0
(1052 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 509 e-144
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 464 e-131
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 461 e-130
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 460 e-129
NP205681 receptor protein kinase-like protein 456 e-128
TC229875 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repea... 381 e-105
TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete 271 1e-72
TC234472 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repea... 251 1e-66
TC209281 similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat recep... 236 5e-62
TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase P... 221 1e-57
TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2,... 205 7e-53
TC231717 weakly similar to UP|Q8VZG8 (Q8VZG8) AT4g08850/T32A17_1... 193 3e-49
TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-l... 191 1e-48
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 186 4e-47
TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial ... 180 3e-45
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 171 2e-42
CO979898 170 3e-42
TC226549 UP|Q84P43 (Q84P43) Protein kinase Pti1, complete 168 1e-41
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 166 5e-41
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 165 9e-41
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 509 bits (1312), Expect = e-144
Identities = 353/1041 (33%), Positives = 544/1041 (51%), Gaps = 11/1041 (1%)
Frame = +3
Query: 2 IMFIILFM-ISWPQAV-AEDSEAQALLKWKHSFDNQSQSLLSTWKNTTNTCTKWKGIFCD 59
++F+ LF +P+ + A SE +ALL + + + +LS+W + C+ W G+ CD
Sbjct: 33 VLFVFLFFHFHFPETLSAPISEYRALLSLRSVITDATPPVLSSWNASIPYCS-WLGVTCD 209
Query: 60 NSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFS 119
N + ++ +NL L GTL S + L L++ N F G IPP + +S + LN S
Sbjct: 210 NRRHVTALNLTGLDLSGTL-SADVAHLPFLSNLSLAANKFSGPIPPSLSALSGLRYLNLS 386
Query: 120 LNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPE 179
N + + P E++ L+SL+ +D ++G +P ++ + NL +L LGGN F G IPPE
Sbjct: 387 NNVFNETFPSELWRLQSLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQ-IPPE 563
Query: 180 IGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSN-NILSGVIPETIGNMSKLNKLY 238
G+ +L +L++ L G+IP EIG LT+L + + N +G IP IGN+S+L +L
Sbjct: 564 YGRWQRLQYLAVSGNELDGTIPPEIGNLTSLRELYIGYYNTYTGGIPPEIGNLSELVRLD 743
Query: 239 LAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIP 298
+A L G IP +L + L ++L +LSGS+ + NL ++ + L N LSG IP
Sbjct: 744 VAY-CALSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIP 920
Query: 299 STIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVF 358
++ G LKN+ L L N+L G+IP IG L L+ + ENNLTG+IP +G RL +
Sbjct: 921 ASFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNLTGSIPEGLGKNGRLNLV 1100
Query: 359 EVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIP 418
++++NKL G LP +CSG L L N GPIP
Sbjct: 1101 DLSSNKL------------------------TGTLPPYLCSGNTLQTLITLGNFLFGPIP 1208
Query: 419 TSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTF 478
SL C S+ RIR+ N + G I + P L ++ DN L G ++NL
Sbjct: 1209 ESLGTCESLTRIRMGENFLNGSIPKGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNL--- 1379
Query: 479 QISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIP 538
G++ LS+NQ +G L +G S+ L L N FT IP
Sbjct: 1380 ---------------------GQITLSNNQLSGALSPSIGNFSSVQKLLLDGNMFTGRIP 1496
Query: 539 TEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFRSSLASLDL 598
T+ G LQ+L +D GN+ SG I E+++ L L+LSRN++ G IP+ ++
Sbjct: 1497 TQIGRLQQLSKIDFSGNKFSGPIAPEISQCKLLTFLDLSRNELSGDIPN---------EI 1649
Query: 599 SGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPS-FSSM-SLDFVNISNNQLEGPLPDN 656
+G R+ L+ LNLS N L G+IPS SSM SL V+ S N L G +P
Sbjct: 1650 TGMRI-------------LNYLNLSKNHLVGSIPSSISSMQSLTSVDFSYNNLSGLVPGT 1790
Query: 657 PAFLHAPFESFKNNKDLCGNFKGLDPCGSRKSKNVLRSVLIALGALILVLFGVGISMYTL 716
F + + SF N DLCG + G C + + + L + + +L VG+ + ++
Sbjct: 1791 GQFSYFNYTSFLGNPDLCGPYLGA--CKGGVANGAHQPHVKGLSSSLKLLLVVGLLLCSI 1964
Query: 717 GRRKKSNEKNQTEEQTQRGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKA 776
+ K ++ ++ + + + ++ F + + + +IG G G VYK
Sbjct: 1965 AFAVAAIFKARSLKKASEARAWKLTAFQ-RLDF-TVDDVLHCLKEDNIIGKGGAGIVYKG 2138
Query: 777 ELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLV 836
+ +G VAVK+L ++ SH F +EI+TL IRHR+I++L GFCS+ + + LV
Sbjct: 2139 AMPNGDHVAVKRLPAMSRGS-SH--DHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLV 2309
Query: 837 YKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVL 896
Y+++ GSLG++L+ + W+ R + A L YLHHDCSP I+HRD+ S N+L
Sbjct: 2310 YEYMPNGSLGEVLHGK-KGGHLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNIL 2486
Query: 897 LNLDYEAQVSDFGTAKFLKPGLLS--WTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLA 954
L+ ++EA V+DFG AKFL+ S + AG++GY APE A T++V+EK DVYSFGV+
Sbjct: 2487 LDSNHEAHVADFGLAKFLQDSGTSECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVL 2666
Query: 955 LEIIVGKHP----GDLISLFLSQSTRLMANNMLLIDVLDQRPQHVMKPVDEEVILIARLA 1010
LE+I G+ P GD + + +N ++ VLD R V P+ EV+ + +A
Sbjct: 2667 LELITGRKPVGEFGDGVDIVQWVRKMTDSNKEGVLKVLDPRLPSV--PL-HEVMHVFYVA 2837
Query: 1011 FACLNQNPRSRPTMDQVSKML 1031
C+ + RPTM +V ++L
Sbjct: 2838 MLCVEEQAVERPTMREVVQIL 2900
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 464 bits (1194), Expect = e-131
Identities = 321/951 (33%), Positives = 482/951 (49%), Gaps = 8/951 (0%)
Frame = +3
Query: 89 LQTLNIYNNYFYGTIPPQIGNISKINTLNFSLNPIDGSIPQEMFTLKSLQNIDFSFCKLS 148
L L++ +N F G IPP + +S + LN S N + + P E+ L++L+ +D ++
Sbjct: 45 LSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRLQNLEVLDLYNNNMT 224
Query: 149 GAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLT 208
G +P ++ + NL +L LGGN F G IPPE G+ +L +L++
Sbjct: 225 GVLPLAVAQMQNLRHLHLGGNFFSGQ-IPPEYGRWQRLQYLAV----------------- 350
Query: 209 NLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMS 268
S N L G IP IGN+S L +LY+ G IP + N+S L +
Sbjct: 351 -------SGNELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCG 509
Query: 269 LSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNL 328
LSG IP ++ L ++ L L N LSG++ +GNLK+L+ + L N LSG IPA G L
Sbjct: 510 LSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGEL 689
Query: 329 INLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKND 388
N+ ++ N L G IP IG L L V ++ N G IP GL +S N
Sbjct: 690 KNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGLGKNGRLNLVDLSSNK 869
Query: 389 FVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVY 448
G LP+ +CSG L L N GPIP SL +C S+ RIR+ N + G I +
Sbjct: 870 LTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMGENFLNGSIPRGLFGL 1049
Query: 449 PNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQ 508
P L ++ DN L G ++NL G++ LS+NQ
Sbjct: 1050 PKLTQVELQDNYLSGEFPEVGSVAVNL------------------------GQITLSNNQ 1157
Query: 509 FTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAEL 568
+G LP +G S+ L L N FT IP + G LQ+L +D GN+ SG I E+++
Sbjct: 1158 LSGVLPPSIGNFSSVQKLILDGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQC 1337
Query: 569 PKLRMLNLSRNKIEGSIPSLFRSSLASLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLS 628
L L+LSRN++ G IP+ +++G R+ L+ LNLS N L
Sbjct: 1338 KLLTFLDLSRNELSGDIPN---------EITGMRI-------------LNYLNLSRNHLV 1451
Query: 629 GTIPS-FSSM-SLDFVNISNNQLEGPLPDNPAFLHAPFESFKNNKDLCGNFKGLDPCGSR 686
G IPS SSM SL V+ S N L G +P F + + SF N DLCG + G C
Sbjct: 1452 GGIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNYTSFLGNPDLCGPYLGA--CKDG 1625
Query: 687 KSKNVLRSVLIALGALILVLFGVGISMYTLGRRKKSNEKNQTEEQTQRGVLFSIWSHDGK 746
+ + + L + +L VG+ + ++ + K ++ ++ + + + +
Sbjct: 1626 VANGAHQPHVKGLSSSFKLLLVVGLLLCSIAFAVAAIFKARSLKKASGARAWKLTAFQ-R 1802
Query: 747 MMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFM 806
+ F + + + +IG G G VYK + +G VAVK+L ++ SH F
Sbjct: 1803 LDF-TVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGS-SH--DHGFN 1970
Query: 807 SEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNV 866
+EI+TL IRHR+I++L GFCS+ + + LVY+++ GSLG++L+ + W+ R +
Sbjct: 1971 AEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGK-KGGHLHWDTRYKI 2147
Query: 867 VKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLS--WTQF 924
A L YLHHDCSP I+HRD+ S N+LL+ ++EA V+DFG AKFL+ S +
Sbjct: 2148 AVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGTSECMSAI 2327
Query: 925 AGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHP----GDLISLFLSQSTRLMAN 980
AG++GY APE A T++V+EK DVYSFGV+ LE+I G+ P GD + + +N
Sbjct: 2328 AGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVGEFGDGVDIVQWVRKMTDSN 2507
Query: 981 NMLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
++ VLD R V P+ EV+ + +A C+ + RPTM +V ++L
Sbjct: 2508 KEGVLKVLDPRLPSV--PL-HEVMHVFYVAMLCVEEQAVERPTMREVVQIL 2651
Score = 230 bits (587), Expect = 2e-60
Identities = 155/475 (32%), Positives = 229/475 (47%), Gaps = 3/475 (0%)
Frame = +3
Query: 67 INLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFSLNPIDGS 126
+NL N T S S NL+ L++YNN G +P + + + L+ N G
Sbjct: 126 LNLSNNVFNETFPS-ELSRLQNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQ 302
Query: 127 IPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKL 186
IP E + LQ + S +L G IP IGNLS+L L +G N IPPEIG L++L
Sbjct: 303 IPPEYGRWQRLQYLAVSGNELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEIGNLSEL 482
Query: 187 WFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLY 246
L C L G IP +G L L + L N LSG + +GN+ L + L+ N L
Sbjct: 483 VRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLS-NNMLS 659
Query: 247 GPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKN 306
G IP + ++TL+ LF L G+IPE + L + + L N +G+IP +G
Sbjct: 660 GEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGLGKNGR 839
Query: 307 LQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLH 366
L + L N+L+G++P + + L + N L G IP ++G+ LT + N L+
Sbjct: 840 LNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMGENFLN 1019
Query: 367 GRIPNGLYNITNWFSFIVSKNDFVGHLP---SQICSGGLLTLLNADHNRFTGPIPTSLKN 423
G IP GL+ + + N G P S + G +TL N N+ +G +P S+ N
Sbjct: 1020GSIPRGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLGQITLSN---NQLSGVLPPSIGN 1190
Query: 424 CSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNN 483
SS++++ L+ N G I G L D S NK G I P + L +S N
Sbjct: 1191FSSVQKLILDGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRN 1370
Query: 484 NISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIP 538
+SG IP E+ G+ L L+LS N G +P + M+SL + S N+ + +P
Sbjct: 1371ELSGDIPNEITGMRILNYLNLSRNHLVGGIPSSISSMQSLTSVDFSYNNLSGLVP 1535
Score = 152 bits (385), Expect = 6e-37
Identities = 106/345 (30%), Positives = 170/345 (48%), Gaps = 5/345 (1%)
Frame = +3
Query: 317 LSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLYNI 376
LSG + A + +L L + S+ N +G IP ++ L+ L ++ N + P+ L +
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELSRL 182
Query: 377 TNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQ 436
N L +L+ +N TG +P ++ ++ + L N
Sbjct: 183 QN------------------------LEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNF 290
Query: 437 IEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQIS-NNNISGVIPLELIG 495
G I ++G + L+Y VS N+L G I P G +L I N +G IP E+
Sbjct: 291 FSGQIPPEYGRWQRLQYLAVSGNELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEIGN 470
Query: 496 LTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGN 555
L++L RL + +G++P LG ++ L L L N + S+ E G L+ L+ +DL N
Sbjct: 471 LSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNN 650
Query: 556 ELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFRS--SLASLDLSGNRLNGKIPEILGF 613
LSG IP EL + +LNL RNK+ G+IP +L + L N G IPE LG
Sbjct: 651 MLSGEIPARFGELKNITLLNLFRNKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGLGK 830
Query: 614 LGQLSMLNLSHNMLSGTIPSF--SSMSLDFVNISNNQLEGPLPDN 656
G+L++++LS N L+GT+P++ S +L + N L GP+P++
Sbjct: 831 NGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPES 965
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 461 bits (1187), Expect = e-130
Identities = 340/1053 (32%), Positives = 525/1053 (49%), Gaps = 22/1053 (2%)
Frame = +1
Query: 2 IMFIILFMISWPQAVAEDSEAQALLKWKHSF--DNQSQSLLSTWKNTTNTCTK--WKGIF 57
++ I F+ + ++ ++LLK K S D L WK + + G+
Sbjct: 115 LLLFIFFIWLRVATCSSFTDMESLLKLKDSMKGDKAKDDALHDWKFFPSLSAHCFFSGVK 294
Query: 58 CDNSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLN 117
CD + IN+ SF L +G +PP+IG + K+ L
Sbjct: 295 CDRELRVVAINV---------------SFVPL----------FGHLPPEIGQLDKLENLT 399
Query: 118 FSLNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIG-NLSNLLYLDLGGNNFVGTPI 176
S N + G +P+E+ L SL++++ S SG P I ++ L LD+ NNF G P+
Sbjct: 400 VSQNNLTGVLPKELAALTSLKHLNISHNVFSGHFPGQIILPMTKLEVLDVYDNNFTG-PL 576
Query: 177 PPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNK 236
P E+ KL KL +L + GSIP+ +L + LS N LSG IP+++ + L
Sbjct: 577 PVELVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGKIPKSLSKLKTLRY 756
Query: 237 LYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGT 296
L L N G IP +M SL + L + +LSG IP S+ NL N++ L L N L+GT
Sbjct: 757 LKLGYNNAYEGGIPPEFGSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLTGT 936
Query: 297 IPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLT 356
IPS + + +L L L +N L+G IP + L NL + +NNL G++P+ +G L L
Sbjct: 937 IPSELSAMVSLMSLDLSINDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLE 1116
Query: 357 VFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGP 416
++ N +P L F V KN F G +P +C G L + N F GP
Sbjct: 1117 TLQLWDNNFSFVLPPNLGQNGKLKFFDVIKNHFTGLIPRDLCKSGRLQTIMITDNFFRGP 1296
Query: 417 IPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLD 476
IP + NC S+ +IR S+N L+G + K ++
Sbjct: 1297 IPNEIGNCKSLTKIR------------------------ASNNYLNGVVPSGIFKLPSVT 1404
Query: 477 TFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDS 536
+++NN +G +P E+ G LG L LS+N F+GK+P L +++L L L N F
Sbjct: 1405 IIELANNRFNGELPPEISG-ESLGILTLSNNLFSGKIPPALKNLRALQTLSLDANEFVGE 1581
Query: 537 IPTEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFR--SSLA 594
IP E L L V+++ GN L+G IP + L ++LSRN +EG IP + + L+
Sbjct: 1582 IPGEVFDLPMLTVVNISGNNLTGPIPTTLTRCVSLTAVDLSRNMLEGKIPKGIKNLTDLS 1761
Query: 595 SLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPSFSSMSL----DFVNISNNQLE 650
++S N+++G +PE + F+ L+ L+LS+N G +P+ ++ F N
Sbjct: 1762 IFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPTGGQFAVFSEKSFAGNPNLCTS 1941
Query: 651 GPLPDNPAFLHAPFESFKNNKDLCGNFKGLDPCGSRKSKNVLRSVLIALGALILVLFGVG 710
P++ + P ++ K + S KS V+ ++IALG L+ V
Sbjct: 1942 HSCPNSSLY---PDDALKKRRG----------PWSLKSTRVI-VIVIALGTAALL---VA 2070
Query: 711 ISMYTLGRRKKSNEKNQTEEQTQRGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQ 770
+++Y + RRK + K QR E+++E + ++ +IG G
Sbjct: 2071 VTVYMMRRRKMNLAKTWKLTAFQR----------LNFKAEDVVECLK---EENIIGKGGA 2211
Query: 771 GNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHS 830
G VY+ + +G VA+K+L S + F +EIETL IRHRNI++L G+ S+
Sbjct: 2212 GIVYRGSMPNGTDVAIKRL----VGAGSGRNDYGFKAEIETLGKIRHRNIMRLLGYVSNK 2379
Query: 831 KFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDI 890
+ + L+Y+++ GSLG+ L+ + WE R + A L YLHHDCSP IIHRD+
Sbjct: 2380 ETNLLLYEYMPNGSLGEWLHG-AKGGHLKWEMRYKIAVEAAKGLCYLHHDCSPLIIHRDV 2556
Query: 891 SSKNVLLNLDYEAQVSDFGTAKFL-KPGL-LSWTQFAGTFGYAAPELAQTMEVNEKCDVY 948
S N+LL+ D EA V+DFG AKFL PG S + AG++GY APE A T++V+EK DVY
Sbjct: 2557 KSNNILLDGDLEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVY 2736
Query: 949 SFGVLALEIIVGKHP----GDLISLF-LSQSTRL----MANNMLLIDVLDQRPQHVMKPV 999
SFGV+ LE+I+G+ P GD + + TRL ++ L++ V+D P+ P+
Sbjct: 2737 SFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTRLELAQPSDAALVLAVVD--PRLSGYPL 2910
Query: 1000 DEEVILIARLAFACLNQNPRSRPTMDQVSKMLA 1032
VI + +A C+ + +RPTM +V ML+
Sbjct: 2911 -TSVIYMFNIAMMCVKEMGPARPTMREVVHMLS 3006
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 460 bits (1183), Expect = e-129
Identities = 334/1054 (31%), Positives = 522/1054 (48%), Gaps = 23/1054 (2%)
Frame = +1
Query: 1 MIMFIILFMISWPQAVAEDSEAQALLKWK-HSFDNQSQSLLSTWKNTTNTCTKWKGIFCD 59
M + ++ F+ A SE +ALL +K S + LS+W ++T C+ W G+ CD
Sbjct: 100 MRVLVLFFLFLHSLQAARISEYRALLSFKASSLTDDPTHALSSWNSSTPFCS-WFGLTCD 276
Query: 60 NSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFS 119
+ + +++ LN +
Sbjct: 277 SRRHVTS-------------------------------------------------LNLT 309
Query: 120 LNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPE 179
+ G++ ++ L L ++ + K SG IP S LS L +L+L N F T P +
Sbjct: 310 SLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIPASFSALSALRFLNLSNNVFNAT-FPSQ 486
Query: 180 IGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYL 239
+ +L L L + N+ G +P + + L + L N SG IP G L L L
Sbjct: 487 LNRLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQIPPEYGTWQHLQYLAL 666
Query: 240 AKNTKLYGPIPHSLWNMSSLTLIYL-FNMSLSGSIPESVENLINVNELALDRNRLSGTIP 298
+ N +L G I L N+SSL +Y+ + + SG IP + NL N+ L LSG IP
Sbjct: 667 SGN-ELAGTIAPELGNLSSLRELYIGYYNTYSGGIPPEIGNLSNLVRLDAAYCGLSGEIP 843
Query: 299 STIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVF 358
+ +G L+NL LFL +N LSGS+ +G+L +L S + N L+G +P + L LT+
Sbjct: 844 AELGKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGEVPASFAELKNLTLL 1023
Query: 359 EVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIP 418
+ NKLHG IP +FVG LP+ L +L N FTG IP
Sbjct: 1024 NLFRNKLHGAIP-----------------EFVGELPA-------LEVLQLWENNFTGSIP 1131
Query: 419 TSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTF 478
+L N + + L N+I G + + L+ N L G I + GK +L+
Sbjct: 1132 QNLGNNGRLTLVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSLGKCKSLNRI 1311
Query: 479 QISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIP 538
++ N ++G IP L GL KL ++ L N TG+ P++ L + LSNN + S+P
Sbjct: 1312 RMGENFLNGSIPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQLSGSLP 1491
Query: 539 TEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSI-PSLFRSSLAS-L 596
+ G ++ L L GNE +G IP ++ L +L ++ S NK G I P + + L + +
Sbjct: 1492 STIGNFTSMQKLLLNGNEFTGRIPPQIGMLQQLSKIDFSHNKFSGPIAPEISKCKLLTFI 1671
Query: 597 DLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPS--FSSMSLDFVNISNNQLEGPLP 654
DLSGN L+G+IP + + L+ LNLS N L G+IP S SL V+ S N G +P
Sbjct: 1672 DLSGNELSGEIPNKITSMRILNYLNLSRNHLDGSIPGNIASMQSLTSVDFSYNNFSGLVP 1851
Query: 655 DNPAFLHAPFESFKNNKDLCGNFKGLDPC------GSRKS--KNVLRS---VLIALGALI 703
F + + SF N +LCG + G PC G R+ K S +L+ +G L+
Sbjct: 1852 GTGQFGYFNYTSFLGNPELCGPYLG--PCKDGVANGPRQPHVKGPFSSSLKLLLVIGLLV 2025
Query: 704 LVLFGVGISMYTLGRRKKSNEKNQTEEQTQRGVLFSIWSHDGKMMFENIIEATENFDDKY 763
+ +++ KK++E + + + F++ +++++ + +
Sbjct: 2026 CSILFAVAAIFKARALKKASEARAWKLTAFQRLDFTV---------DDVLDCLK---EDN 2169
Query: 764 LIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKL 823
+IG G G VYK + +G VAVK+L ++ SH F +EI+TL IRHR+I++L
Sbjct: 2170 IIGKGGAGIVYKGAMPNGDNVAVKRLPAMSRGS-SH--DHGFNAEIQTLGRIRHRHIVRL 2340
Query: 824 HGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSP 883
GFCS+ + + LVY+++ GSLG++L+ + W+ R + A L YLHHDCSP
Sbjct: 2341 LGFCSNHETNLLVYEYMPNGSLGEVLHGK-KGGHLHWDTRYKIAVEAAKGLCYLHHDCSP 2517
Query: 884 PIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLS--WTQFAGTFGYAAPELAQTMEV 941
I+HRD+ S N+LL+ ++EA V+DFG AKFL+ S + AG++GY APE A T++V
Sbjct: 2518 LIVHRDVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYTLKV 2697
Query: 942 NEKCDVYSFGVLALEIIVGKHP----GDLISLFLSQSTRLMANNMLLIDVLDQRPQHVMK 997
+EK DVYSFGV+ LE++ G+ P GD + + +N ++ VLD R V
Sbjct: 2698 DEKSDVYSFGVVLLELVTGRKPVGEFGDGVDIVQWVRKMTDSNKEGVLKVLDSRLPSV-- 2871
Query: 998 PVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
P+ EV+ + +A C+ + RPTM +V ++L
Sbjct: 2872 PL-HEVMHVFYVAMLCVEEQAVERPTMREVVQIL 2970
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 456 bits (1174), Expect = e-128
Identities = 331/1058 (31%), Positives = 511/1058 (48%), Gaps = 27/1058 (2%)
Frame = +1
Query: 2 IMFIILFMISWPQAVAEDSEAQALLKWKHSF--DNQSQSLLSTWKNTTNTCTK--WKGIF 57
++ + F+ + S+ ALLK K S D L WK +T+ + G+
Sbjct: 25 LLLFVFFIWLHVATCSSFSDMDALLKLKESMKGDRAKDDALHDWKFSTSLSAHCFFSGVS 204
Query: 58 CDNSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLN 117
CD + IN+ SF L +G +PP+IG + K+ L
Sbjct: 205 CDQELRVVAINV---------------SFVPL----------FGHVPPEIGELDKLENLT 309
Query: 118 FSLNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIG-NLSNLLYLDLGGNNFVGTPI 176
S N + G +P+E+ L SL++++ S SG P I ++ L LD+ NNF G+ +
Sbjct: 310 ISQNNLTGELPKELAALTSLKHLNISHNVFSGYFPGKIILPMTELEVLDVYDNNFTGS-L 486
Query: 177 PPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNK 236
P E KL KL +L + GSIP+ +L + LS N LSG IP+++ + L
Sbjct: 487 PEEFVKLEKLKYLKLDGNYFSGSIPESYSEFKSLEFLSLSTNSLSGNIPKSLSKLKTLRI 666
Query: 237 LYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGT 296
L L N G IP M SL + L + +LSG IP S+ N+ N++ L L N L+GT
Sbjct: 667 LKLGYNNAYEGGIPPEFGTMESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLTGT 846
Query: 297 IPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLT 356
IPS + ++ +L L L N L+G IP L NL + NNL G++P+ +G L L
Sbjct: 847 IPSELSDMVSLMSLDLSFNGLTGEIPTRFSQLKNLTLMNFFHNNLRGSVPSFVGELPNLE 1026
Query: 357 VFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGP 416
++ N +P L + F V+KN F G +P +C G L N F GP
Sbjct: 1027 TLQLWENNFSSELPQNLGQNGKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGP 1206
Query: 417 IPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLD 476
IP + NC S+ +IR S+N L+G + K ++
Sbjct: 1207 IPNEIANCKSLTKIR------------------------ASNNYLNGAVPSGIFKLPSVT 1314
Query: 477 TFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDS 536
+++NN +G +P E+ G LG L LS+N FTGK+P L +++L L L N F
Sbjct: 1315 IIELANNRFNGELPPEISG-DSLGILTLSNNLFTGKIPPALKNLRALQTLSLDTNEFLGE 1491
Query: 537 IPTEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFR--SSLA 594
IP E L L V+++ GN L+G IP L ++LSRN ++G IP + + L+
Sbjct: 1492 IPGEVFDLPMLTVVNISGNNLTGPIPTTFTRCVSLAAVDLSRNMLDGEIPKGMKNLTDLS 1671
Query: 595 SLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPSFSSMSLDFVNISNNQLEGPLP 654
++S N+++G +P+ + F+ L+ L+LS+N G +P+
Sbjct: 1672 IFNVSINQISGSVPDEIRFMLSLTTLDLSYNNFIGKVPT--------------------- 1788
Query: 655 DNPAFLHAPFESFKNNKDLCGNF--------KGLDPCGSRKSKNVLRSVLIALGALILVL 706
FL +SF N +LC + K P + ++ ++ + +A A++
Sbjct: 1789 -GGQFLVFSDKSFAGNPNLCSSHSCPNSSLKKRRGPWSLKSTRVIVMVIALATAAIL--- 1956
Query: 707 FGVGISMYTLGRRKKSNEKNQTEEQTQRGVLFSIWSHDG-KMMFENIIEATENFDDKYLI 765
V + Y RRK L W G + + E E ++ +I
Sbjct: 1957 --VAGTEYMRRRRKLK--------------LAMTWKLTGFQRLNLKAEEVVECLKEENII 2088
Query: 766 GVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHG 825
G G G VY+ + +G VA+K+L S + F +EIET+ IRHRNI++L G
Sbjct: 2089 GKGGAGIVYRGSMRNGSDVAIKRL----VGAGSGRNDYGFKAEIETVGKIRHRNIMRLLG 2256
Query: 826 FCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPI 885
+ S+ + + L+Y+++ GSLG+ L+ + WE R + A L YLHHDCSP I
Sbjct: 2257 YVSNKETNLLLYEYMPNGSLGEWLHG-AKGGHLKWEMRYKIAVEAAKGLCYLHHDCSPLI 2433
Query: 886 IHRDISSKNVLLNLDYEAQVSDFGTAKFLKP--GLLSWTQFAGTFGYAAPELAQTMEVNE 943
IHRD+ S N+LL+ +EA V+DFG AKFL S + AG++GY APE A T++V+E
Sbjct: 2434 IHRDVKSNNILLDAHFEAHVADFGLAKFLYDLGSSQSMSSIAGSYGYIAPEYAYTLKVDE 2613
Query: 944 KCDVYSFGVLALEIIVGKHP----GDLISLF-LSQSTRL----MANNMLLIDVLDQRPQH 994
K DVYSFGV+ LE+I+G+ P GD + + TRL ++ +++ V+D P+
Sbjct: 2614 KSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTRLELSQPSDAAVVLAVVD--PRL 2787
Query: 995 VMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKMLA 1032
P+ VI + +A C+ + +RPTM +V ML+
Sbjct: 2788 SGYPL-ISVIYMFNIAMMCVKEVGPTRPTMREVVHMLS 2898
>TC229875 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat
receptor-like protein kinase 1, partial (23%)
Length = 1141
Score = 381 bits (978), Expect = e-105
Identities = 192/298 (64%), Positives = 231/298 (77%), Gaps = 4/298 (1%)
Frame = +2
Query: 743 HDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSS 802
H GKM+FENIIEATE+FDDK+LIGVG QG VYKA L +G VVAVKKLH + + E+ +
Sbjct: 2 HQGKMVFENIIEATEDFDDKHLIGVGGQGCVYKAVLPTGQVVAVKKLHSVPNGEMLNL-- 175
Query: 803 KSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEK 862
K+F EI+ L+ IRHRNI+KL+GFCSHS+FSFLV +FLE GS+G+ L D QA AFDW K
Sbjct: 176 KAFTCEIQALTEIRHRNIVKLYGFCSHSQFSFLVCEFLENGSVGKTLKDDGQAMAFDWYK 355
Query: 863 RVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSWT 922
RVNVVK VANAL Y+HH+CSP I+HRDISSKNVLL+ +Y A VSDFGTAKFL P +WT
Sbjct: 356 RVNVVKDVANALCYMHHECSPRIVHRDISSKNVLLDSEYVAHVSDFGTAKFLNPDSSNWT 535
Query: 923 QFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLMA--- 979
F GTFGYAAPELA TMEVNEKCDVYSFGVLA EI++GKHPGD+IS L S +
Sbjct: 536 SFVGTFGYAAPELAYTMEVNEKCDVYSFGVLAWEILIGKHPGDVISCLLGSSPSTLVAST 715
Query: 980 -NNMLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKMLAIGKS 1036
++M L+D LDQR H KP+ +EV IA++A ACL ++PRSRPTM+QV+ L + S
Sbjct: 716 LDHMALMDKLDQRLPHPTKPIGKEVASIAKIAMACLTESPRSRPTMEQVANELVMSSS 889
>TC204550 UP|Q8L3Y5 (Q8L3Y5) Receptor-like kinase RHG1, complete
Length = 2659
Score = 271 bits (692), Expect = 1e-72
Identities = 224/775 (28%), Positives = 349/775 (44%), Gaps = 36/775 (4%)
Frame = +2
Query: 293 LSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNL 352
L G I IG L+ L+ L L N++ GSIP+T+G L NL + N LTG+IP ++G
Sbjct: 473 LRGRITDKIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFC 652
Query: 353 NRLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNR 412
L +++ N L G IP L N T + +S N F G LP+ + LT L+ +N
Sbjct: 653 PLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLTFLSLQNNN 832
Query: 413 FTGPIPTSLKNCSS-----IERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISP 467
+G +P S S ++ + L+ N GD+ G L +S NK G I
Sbjct: 833 LSGSLPNSWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAIPN 1012
Query: 468 NWGKSLNLDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLK 527
G L T ISNN ++G +P L L+ L L+ +N ++P+ LG +++L L
Sbjct: 1013 EIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLI 1192
Query: 528 LSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPS 587
LS N F SG IP+ +A + LR L+LS N G IP
Sbjct: 1193 LSRNQF------------------------SGHIPSSIANISSLRQLDLSLNNFSGEIPV 1300
Query: 588 LFRS--SLASLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPSFSSMSLDFVNIS 645
F S SL ++S N L+G +P +L F+S S +
Sbjct: 1301 SFDSQRSLNLFNVSYNSLSGSVPPLLA-------------------KKFNSSSF----VG 1411
Query: 646 NNQLEGPLPDNPAFLHAPFESFKNNKDLCGNFKGLDPCGSRKSKNVLRSVLIALGALILV 705
N QL G P P F AP ++ + + + + +L+ G L++V
Sbjct: 1412 NIQLCGYSPSTPCFSQAP------SQGVIAPPPEVSKHHHHRKLSTKDIILVVAGVLLVV 1573
Query: 706 LFGV-GISMYTLGRRKKSNEKNQTEEQTQRGVLF--------SIW--------------- 741
L + + ++ L R++ +++ + R V + S W
Sbjct: 1574 LIILCCVLLFCLIRKRSTSKAGNGQATEGRAVTYEDRKRSPSSCWWLMLKQVGRLEGNLV 1753
Query: 742 SHDGKMMF--ENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISH 799
DG M F ++++ AT ++G + G VYKA L G VAVK+L E+I+
Sbjct: 1754 HFDGPMAFTADDLLCATAE-----IMGKSTNGTVYKAILEDGSQVAVKRLR----EKIAK 1906
Query: 800 FSSKSFMSEIETLSGIRHRNIIKLHGFCSHSK-FSFLVYKFLEGGSLGQMLNSDTQATAF 858
+ F SE+ L IRH N++ L + K LV+ ++ GSL L+ T
Sbjct: 1907 -GHREFESEVSVLGKIRHPNVLALRAYYLGPKGEKLLVFDYMSKGSLASFLHGGGTETFI 2083
Query: 859 DWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGL 918
DW R+ + + +A L LH IIH +++S NVLL+ + A+++DFG ++ +
Sbjct: 2084 DWPTRMKIAQDLARGLFCLH--SQENIIHGNLTSSNVLLDENTNAKIADFGLSRLMSTTA 2257
Query: 919 LS-WTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPG-DLISLFLSQSTR 976
S AG GY APEL++ + N K D+YS GV+ LE++ K PG + L L Q
Sbjct: 2258 NSNVIATAGALGYRAPELSKLKKANTKTDIYSLGVILLELLTRKSPGVSMNGLDLPQWVA 2437
Query: 977 LMANNMLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
+ +V D V +E++ +LA C++ +P +RP + QV + L
Sbjct: 2438 SVVKEEWTNEVFDADLMRDASTVGDELLNTLKLALHCVDPSPSARPEVHQVLQQL 2602
Score = 182 bits (461), Expect = 9e-46
Identities = 121/349 (34%), Positives = 171/349 (48%), Gaps = 5/349 (1%)
Frame = +2
Query: 147 LSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGF 206
L G I + IG L L L L N +G IP +G L L + + L GSIP +GF
Sbjct: 473 LRGRITDKIGQLQGLRKLSLHDNQ-IGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGF 649
Query: 207 LTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFN 266
L +DLSNN+L+G IP ++ N +KL L L+ N+ GP+P SL + SLT + L N
Sbjct: 650 CPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNS-FSGPLPASLTHSFSLTFLSLQN 826
Query: 267 MSLSGSIPES-----VENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSI 321
+LSGS+P S + L LD N +G +P+++G+L+ L + L N+ SG+I
Sbjct: 827 NNLSGSLPNSWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAI 1006
Query: 322 PATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLYNITNWFS 381
P IG L L + + N L G +P T+ NL+ LT+ N L +IP L + N
Sbjct: 1007PNEIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSV 1186
Query: 382 FIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDI 441
I+S+N F GH+PS I + L L+ N F+G IP S
Sbjct: 1187LILSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVS--------------------- 1303
Query: 442 AQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGVIP 490
F +L F+VS N L G + P K N +F + N + G P
Sbjct: 1304---FDSQRSLNLFNVSYNSLSGSVPPLLAKKFNSSSF-VGNIQLCGYSP 1438
Score = 179 bits (454), Expect = 6e-45
Identities = 125/395 (31%), Positives = 201/395 (50%), Gaps = 8/395 (2%)
Frame = +2
Query: 12 WPQAVAEDSEAQALLKWKHSFDNQSQSLLSTWKNTT-NTCTK-WKGIFCDNSKSISTINL 69
W V S AL +K + + L +W ++ C+ W GI C + I I L
Sbjct: 287 WDGVVVTASNLLALEAFKQELADP-EGFLRSWNDSGYGACSGGWVGIKCAQGQVI-VIQL 460
Query: 70 ENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFSLNPIDGSIPQ 129
GL+G + L+ L++++N G+IP +G + + + N + GSIP
Sbjct: 461 PWKGLRGRITD-KIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPL 637
Query: 130 EMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWFL 189
+ LQ++D S L+GAIP S+ N + L +L+L N+F G P+P + L FL
Sbjct: 638 SLGFCPLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSG-PLPASLTHSFSLTFL 814
Query: 190 SIQKCNLIGSIP------KEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNT 243
S+Q NL GS+P + GF LI L +N +G +P ++G++ +LN++ L+ N
Sbjct: 815 SLQNNNLSGSLPNSWGGNSKNGFFRLQNLI-LDHNFFTGDVPASLGSLRELNEISLSHN- 988
Query: 244 KLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGN 303
K G IP+ + +S L + + N +L+G++P ++ NL ++ L + N L IP ++G
Sbjct: 989 KFSGAIPNEIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGR 1168
Query: 304 LKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAAN 363
L+NL L L N+ SG IP++I N+ +L + NN +G IP + + L +F V+ N
Sbjct: 1169LRNLSVLILSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVSFDSQRSLNLFNVSYN 1348
Query: 364 KLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQIC 398
L G +P L N SF V G+ PS C
Sbjct: 1349SLSGSVPPLLAKKFNSSSF-VGNIQLCGYSPSTPC 1450
Score = 159 bits (403), Expect = 5e-39
Identities = 100/304 (32%), Positives = 152/304 (49%), Gaps = 5/304 (1%)
Frame = +2
Query: 269 LSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNL 328
L G I + + L + +L+L N++ G+IPST+G L NL+ + L NRL+GSIP ++G
Sbjct: 473 LRGRITDKIGQLQGLRKLSLHDNQIGGSIPSTLGLLPNLRGVQLFNNRLTGSIPLSLGFC 652
Query: 329 INLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKND 388
L S + N LTG IP ++ N +L ++ N G +P L + + + N+
Sbjct: 653 PLLQSLDLSNNLLTGAIPYSLANSTKLYWLNLSFNSFSGPLPASLTHSFSLTFLSLQNNN 832
Query: 389 FVGHLPSQI---CSGGLLTLLNA--DHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQ 443
G LP+ G L N DHN FTG +P SL + + I L N+ G I
Sbjct: 833 LSGSLPNSWGGNSKNGFFRLQNLILDHNFFTGDVPASLGSLRELNEISLSHNKFSGAIPN 1012
Query: 444 DFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGVIPLELIGLTKLGRLH 503
+ G L+ D+S+N L+G++ +L NN + IP L L L L
Sbjct: 1013EIGTLSRLKTLDISNNALNGNLPATLSNLSSLTLLNAENNLLDNQIPQSLGRLRNLSVLI 1192
Query: 504 LSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPN 563
LS NQF+G +P + + SL L LS N+F+ IP F + L + ++ N LSG +P
Sbjct: 1193LSRNQFSGHIPSSIANISSLRQLDLSLNNFSGEIPVSFDSQRSLNLFNVSYNSLSGSVPP 1372
Query: 564 EVAE 567
+A+
Sbjct: 1373LLAK 1384
>TC234472 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat
receptor-like protein kinase 1, partial (16%)
Length = 553
Score = 251 bits (641), Expect = 1e-66
Identities = 115/178 (64%), Positives = 152/178 (84%)
Frame = +1
Query: 737 LFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEE 796
LF+IWS DGK+++ENI+EATE+FD+K+LIGVG QG+VYKA+L +G ++AVKKLH++ + E
Sbjct: 22 LFAIWSFDGKLVYENIVEATEDFDNKHLIGVGGQGSVYKAKLHTGQILAVKKLHLVQNGE 201
Query: 797 ISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQAT 856
+S+ K+F SEI+ L IRHRNI+KL+GFCSHS+ SFLVY+FLE GS+ ++L D QA
Sbjct: 202 LSNI--KAFTSEIQALINIRHRNIVKLYGFCSHSQSSFLVYEFLEKGSIDKILKDDEQAI 375
Query: 857 AFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFL 914
AFDW+ R+N +KGVANALSY+HHDCSPPI+HRDISSKN++L+L+Y A VSDFG A+ L
Sbjct: 376 AFDWDPRINAIKGVANALSYMHHDCSPPIVHRDISSKNIVLDLEYVAHVSDFGAARLL 549
>TC209281 similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat receptor-like
protein kinase 1, partial (14%)
Length = 586
Score = 236 bits (601), Expect = 5e-62
Identities = 117/193 (60%), Positives = 145/193 (74%), Gaps = 3/193 (1%)
Frame = +2
Query: 838 KFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLL 897
+FLE G + ++L D QA A DW KRV++VKGVANAL Y+HHDCSPPI+HRDISSKNVLL
Sbjct: 8 EFLEKGDVKKILKDDEQAIALDWNKRVDIVKGVANALCYMHHDCSPPIVHRDISSKNVLL 187
Query: 898 NLDYEAQVSDFGTAKFLKPGLLSWTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEI 957
+ D A V+DFGTAKFL P +WT FAGT+GYAAPELA TME NEKCDVYSFGV ALEI
Sbjct: 188 DSDDVAHVADFGTAKFLNPDSSNWTSFAGTYGYAAPELAYTMEANEKCDVYSFGVFALEI 367
Query: 958 IVGKHPGDLISLFLSQSTRLMA---NNMLLIDVLDQRPQHVMKPVDEEVILIARLAFACL 1014
+ G+HPGD+ S L S+ M ++M L+ LD+R H P+D+EVI I ++A A L
Sbjct: 368 LFGEHPGDVTSSLLLSSSSTMTSTLDHMSLMVKLDERLPHPTSPIDKEVISIVKIAIARL 547
Query: 1015 NQNPRSRPTMDQV 1027
++ RSRP+M+QV
Sbjct: 548 TESTRSRPSMEQV 586
>TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase PERK1-like,
partial (80%)
Length = 1604
Score = 221 bits (564), Expect = 1e-57
Identities = 140/412 (33%), Positives = 220/412 (52%), Gaps = 17/412 (4%)
Frame = +1
Query: 637 MSLDFVNISNNQLEGPLPDNPAFLHAPFESFKNNKDLCGNFKGLDPCGSRKSKNVLRSVL 696
+SL +N+S N+L G +P + F P +SF N LCGN+ L G+R S+ V S
Sbjct: 1 LSLSLLNVSYNKLFGVIPTSNNFTRFPPDSFIGNPGLCGNWLNLPCHGARPSERVTLSKA 180
Query: 697 ----IALGALILVLFGVGISMYTLGRRKKSNEKNQTEEQTQRGVLFS------IWSHDGK 746
I LGAL+++L M + + + + + + FS + +
Sbjct: 181 AILGITLGALVILL------MVLVAACRPHSPSPFPDGSFDKPINFSPPKLVILHMNMAL 342
Query: 747 MMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSS--KS 804
++E+I+ TEN +KY+IG G+ VYK L + VA+K+++ SH+ K
Sbjct: 343 HVYEDIMRMTENLSEKYIIGYGASSTVYKCVLKNCKPVAIKRIY-------SHYPQCIKE 501
Query: 805 FMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRV 864
F +E+ET+ I+HRN++ L G+ L Y ++E GSL +L+ T+ DWE R+
Sbjct: 502 FETELETVGSIKHRNLVSLQGYSLSPYGHLLFYDYMENGSLWDLLHGPTKKKKLDWELRL 681
Query: 865 NVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLL-SWTQ 923
+ G A L+YLHHDC P IIHRD+ S N+LL+ D+E ++DFG AK L P + T
Sbjct: 682 KIALGAAQGLAYLHHDCCPRIIHRDVKSSNILLDADFEPHLTDFGIAKSLCPSKSHTSTY 861
Query: 924 FAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGK----HPGDLISLFLSQSTRLMA 979
GT GY PE A+T + EK DVYS+G++ LE++ G+ + +L L LS++ A
Sbjct: 862 IMGTIGYIDPEYARTSHLTEKSDVYSYGIVLLELLTGRKAVDNESNLHHLILSKA----A 1029
Query: 980 NNMLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
N ++ + +D K + V + +LA C + P RPTM +V+++L
Sbjct: 1030 TNAVM-ETVDPDITATCKDLG-AVKKVYQLALLCTKRQPADRPTMHEVTRVL 1179
>TC231346 weakly similar to UP|Q6QM04 (Q6QM04) LRR protein WM1.2, partial
(14%)
Length = 1790
Score = 205 bits (522), Expect = 7e-53
Identities = 178/587 (30%), Positives = 281/587 (47%), Gaps = 59/587 (10%)
Frame = +1
Query: 125 GSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLN 184
G IPQ + +L++++N+D +LSG +P+S+G L +L LDL N F PIP L+
Sbjct: 25 GKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFT-CPIPSPFANLS 201
Query: 185 KLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTK 244
L L++ L G+IPK FL NL +++L N L+G +P T+G +S L L L+ N
Sbjct: 202 SLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLSSNL- 378
Query: 245 LYGPIPHS----LWNMSSLTL---------------------IYLFNMSLSGSIPESVEN 279
L G I S L+ + L L + L + + PE ++
Sbjct: 379 LEGSIKESNFVKLFTLKELRLSWTNLFLSVNSGWAPPFQLEYVLLSSFGIGPKFPEWLKR 558
Query: 280 LINVNELALDRNRLSGTIPS--TIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQ 337
+V L + + ++ +PS I L+ +++L L N L G + +N ++
Sbjct: 559 QSSVKVLTMSKAGIADLVPSWFWIWTLQ-IEFLDLSNNLLRGDLS---NIFLNSSVINLS 726
Query: 338 ENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPN---GLYNITNWFSFI-VSKNDFVGHL 393
N G +P+ N+ V VA N + G I G N TN S + S N G L
Sbjct: 727 SNLFKGRLPSVSANVE---VLNVANNSISGTISPFLCGKPNATNKLSVLDFSNNVLSGDL 897
Query: 394 PSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRY 453
L +N N +G IP S+ S +E + L+ N+ G I +++
Sbjct: 898 GHCWVHWQALVHVNLGSNNMSGEIPNSMGYLSQLESLLLDDNRFSGYIPSTLQNCSTMKF 1077
Query: 454 FDVSDNKLHGHISPNWGKSLN-LDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGK 512
D+ +N+L I P+W + L ++ +NN +G I ++ L+ L L +N +G
Sbjct: 1078IDMGNNQLSDTI-PDWMWEMQYLMVLRLRSNNFNGSITQKMCQLSSLIVLDHGNNSLSGS 1254
Query: 513 LPKELGGMKSL--------------FDLKLSNNHFTDS---IPTEFGLLQR-----LEVL 550
+P L MK++ + S NH+ ++ +P + L R + ++
Sbjct: 1255IPNCLDDMKTMAGEDDFFANPSSYSYGSDFSYNHYKETLVLVPKKDELEYRDNLILVRMI 1434
Query: 551 DLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLF--RSSLASLDLSGNRLNGKIP 608
DL N+LSG IP+E+++L LR LNLSRN + G IP+ L SLDLS N ++G+IP
Sbjct: 1435DLSSNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKMKLLESLDLSLNNISGQIP 1614
Query: 609 EILGFLGQLSMLNLSHNMLSGTIPSFSSMSLDFVNIS---NNQLEGP 652
+ L L LS LNLS++ LSG IP+ S+ F +S N +L GP
Sbjct: 1615QSLSDLSFLSFLNLSYHNLSGRIPT-STQLQSFEELSYTGNPELCGP 1752
Score = 130 bits (327), Expect = 3e-30
Identities = 101/337 (29%), Positives = 166/337 (48%), Gaps = 31/337 (9%)
Frame = +1
Query: 65 STINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQI----GNISKINTLNFSL 120
S INL + KG L S++ +N++ LN+ NN GTI P + +K++ L+FS
Sbjct: 709 SVINLSSNLFKGRLPSVS----ANVEVLNVANNSISGTISPFLCGKPNATNKLSVLDFSN 876
Query: 121 NPIDGS------------------------IPQEMFTLKSLQNIDFSFCKLSGAIPNSIG 156
N + G IP M L L+++ + SG IP+++
Sbjct: 877 NVLSGDLGHCWVHWQALVHVNLGSNNMSGEIPNSMGYLSQLESLLLDDNRFSGYIPSTLQ 1056
Query: 157 NLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLS 216
N S + ++D+G N T IP + ++ L L ++ N GSI +++ L++L ++D
Sbjct: 1057 NCSTMKFIDMGNNQLSDT-IPDWMWEMQYLMVLRLRSNNFNGSITQKMCQLSSLIVLDHG 1233
Query: 217 NNILSGVIPETIGNMSKL---NKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSI 273
NN LSG IP + +M + + + ++ YG +N TL+ +
Sbjct: 1234 NNSLSGSIPNCLDDMKTMAGEDDFFANPSSYSYGS--DFSYNHYKETLVLVPKKDEL--- 1398
Query: 274 PESVENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDS 333
E +NLI V + L N+LSG IPS I L L++L L N LSG IP +G + L+S
Sbjct: 1399 -EYRDNLILVRMIDLSSNKLSGAIPSEISKLSALRFLNLSRNHLSGEIPNDMGKMKLLES 1575
Query: 334 FSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIP 370
+ NN++G IP ++ +L+ L+ ++ + L GRIP
Sbjct: 1576 LDLSLNNISGQIPQSLSDLSFLSFLNLSYHNLSGRIP 1686
Score = 92.8 bits (229), Expect = 7e-19
Identities = 50/133 (37%), Positives = 76/133 (56%), Gaps = 2/133 (1%)
Frame = +1
Query: 509 FTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAEL 568
+ GK+P+ + ++++ +L L NN + +P G L+ LEVLDL N + IP+ A L
Sbjct: 19 YRGKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTCPIPSPFANL 198
Query: 569 PKLRMLNLSRNKIEGSIPSLFR--SSLASLDLSGNRLNGKIPEILGFLGQLSMLNLSHNM 626
LR LNL+ N++ G+IP F +L L+L N L G +P LG L L L+LS N+
Sbjct: 199 SSLRTLNLAHNRLNGTIPKSFEFLKNLQVLNLGANSLTGDVPVTLGTLSNLVTLDLSSNL 378
Query: 627 LSGTIPSFSSMSL 639
L G+I + + L
Sbjct: 379 LEGSIKESNFVKL 417
Score = 29.3 bits (64), Expect = 9.5
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 3/49 (6%)
Frame = +1
Query: 629 GTIPSFSS--MSLDFVNISNNQLEGPLPDNPAFL-HAPFESFKNNKDLC 674
G IP S ++ +++ NNQL GPLPD+ L H NN C
Sbjct: 25 GKIPQIISSLQNIKNLDLQNNQLSGPLPDSLGQLKHLEVLDLSNNTFTC 171
>TC231717 weakly similar to UP|Q8VZG8 (Q8VZG8) AT4g08850/T32A17_160, partial
(11%)
Length = 453
Score = 193 bits (491), Expect = 3e-49
Identities = 91/144 (63%), Positives = 117/144 (81%)
Frame = +3
Query: 773 VYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKF 832
VYKA+L +G +VAVKKLH +EE SK+F +E++ L+ I+HRNI+K G+C H +F
Sbjct: 6 VYKAKLPAGQIVAVKKLHAAPNEETP--DSKAFSTEVKALAEIKHRNIVKSLGYCLHPRF 179
Query: 833 SFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISS 892
SFL+Y+FLEGGSL ++L DT+AT FDWE+RV VVKGVA+AL ++HH C PPI+HRDISS
Sbjct: 180 SFLIYEFLEGGSLDKVLTDDTRATMFDWERRVKVVKGVASALYHMHHGCFPPIVHRDISS 359
Query: 893 KNVLLNLDYEAQVSDFGTAKFLKP 916
KNVL++LDYEA +SDFGTAK LKP
Sbjct: 360 KNVLIDLDYEAHISDFGTAKILKP 431
>TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-like, partial
(31%)
Length = 1329
Score = 191 bits (485), Expect = 1e-48
Identities = 144/438 (32%), Positives = 203/438 (45%), Gaps = 10/438 (2%)
Frame = +1
Query: 298 PSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTV 357
PS + N++ L L L N +GSIP + +L NL SV N+++GT+P I L L
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 358 FEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPI 417
+ NK G +P L + S ND VG++P IC G L L N+FTG +
Sbjct: 181 LLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGGL 360
Query: 418 PTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDT 477
+S+ NCSS+ R+RLE N G+I F + P++ Y D+S N G I + ++ L+
Sbjct: 361 -SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEY 537
Query: 478 FQIS-NNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDS 536
F +S N + G+IP + L +L SS + LP
Sbjct: 538 FNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPL--------------------- 654
Query: 537 IPTEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFRS--SLA 594
F + + V+DL N LSG IPN V++ L +NLS N + G IP S L
Sbjct: 655 ----FESCKSISVIDLDSNSLSGTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLG 822
Query: 595 SLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPSFSSMSLDFVNISNNQLEGPLP 654
+DLS N+ NG IP G L +LN+S N +SG+IP+ S L
Sbjct: 823 VVDLSNNKFNGPIPAKFGSSSNLQLLNVSFNNISGSIPTGKSFKL--------------- 957
Query: 655 DNPAFLHAPFESFKNNKDLCGNFKGLDPC-------GSRKSKNVLRSVLIALGALILVLF 707
+F N +LCG L PC GS+ S V R VL+++G L++VL
Sbjct: 958 -------MGRSAFVGNSELCG--APLQPCPDSVGILGSKCSWKVTRIVLLSVG-LLIVLL 1107
Query: 708 GVGISMYTLGRRKKSNEK 725
G+ M L R KS K
Sbjct: 1108GLAFGMSYLRRGIKSQWK 1161
Score = 138 bits (348), Expect = 1e-32
Identities = 99/316 (31%), Positives = 149/316 (46%), Gaps = 2/316 (0%)
Frame = +1
Query: 177 PPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNK 236
P E+ + L L + GSIP+ L NL L+ + N +SG +PE I + L
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 237 LYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGT 296
L L N K G +P SL S L + L G+IP + + +L L N+ +G
Sbjct: 181 L-LIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGG 357
Query: 297 IPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLT 356
+ S+I N +L L L N SG I L ++ + NN G IP+ I +L
Sbjct: 358 L-SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLE 534
Query: 357 VFEVAAN-KLHGRIPNGLYNITNWFSFIVSKNDFVGHLP-SQICSGGLLTLLNADHNRFT 414
F V+ N +L G IP+ +++ +F S LP + C +++++ D N +
Sbjct: 535 YFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPLFESCKS--ISVIDLDSNSLS 708
Query: 415 GPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLN 474
G IP + C ++E+I L N + G I + P L D+S+NK +G I +G S N
Sbjct: 709 GTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSN 888
Query: 475 LDTFQISNNNISGVIP 490
L +S NNISG IP
Sbjct: 889 LQLLNVSFNNISGSIP 936
Score = 133 bits (335), Expect = 4e-31
Identities = 107/339 (31%), Positives = 164/339 (47%), Gaps = 29/339 (8%)
Frame = +1
Query: 250 PHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQY 309
P L N+ LT + L + +GSIPES +L N+ L++ N +SGT+P I L +L+
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 310 LFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTI---GNLNRLTVFEVAANKLH 366
L + N+ SGS+P ++G L N+L G IP I G L +L +F +NK
Sbjct: 181 LLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILF---SNKFT 351
Query: 367 G--------------RIPNGLYN--ITNWFSFI-------VSKNDFVGHLPSQICSGGLL 403
G R+ + L++ IT FS + +S+N+FVG +PS I L
Sbjct: 352 GGLSSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQL 531
Query: 404 TLLNADHN-RFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLH 462
N +N + G IP+ + ++ I D+ F ++ D+ N L
Sbjct: 532 EYFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPL-FESCKSISVIDLDSNSLS 708
Query: 463 GHISPNWGKSLNLDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKS 522
G I K L+ +SNNN++G IP EL + LG + LS+N+F G +P + G +
Sbjct: 709 GTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSN 888
Query: 523 LFDLKLSNNHFTDSIPT--EFGLLQRLEVLDLGGNELSG 559
L L +S N+ + SIPT F L+ R +G +EL G
Sbjct: 889 LQLLNVSFNNISGSIPTGKSFKLMGRSAF--VGNSELCG 999
Score = 126 bits (317), Expect = 4e-29
Identities = 89/310 (28%), Positives = 147/310 (46%), Gaps = 46/310 (14%)
Frame = +1
Query: 84 SSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFSLNPIDGSIPQEMFTLKSLQNIDFS 143
S+ L L++ +N+F G+IP ++ + L+ N + G++P+ + L SL+ +
Sbjct: 13 SNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLETLLIW 192
Query: 144 FCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEI---GKLNKLWFLS---------I 191
K SG++P S+G S L ++D N+ VG IPP+I G+L KL S I
Sbjct: 193 NNKFSGSLPRSLGRNSKLKWVDASTNDLVGN-IPPDICVSGELFKLILFSNKFTGGLSSI 369
Query: 192 QKCNLI-----------GSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLA 240
C+ + G I + L ++ +DLS N G IP I ++L ++
Sbjct: 370 SNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEYFNVS 549
Query: 241 KNTKLYGPIPHSLWNM-----------------------SSLTLIYLFNMSLSGSIPESV 277
N +L G IP W++ S+++I L + SLSG+IP V
Sbjct: 550 YNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPLFESCKSISVIDLDSNSLSGTIPNGV 729
Query: 278 ENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQ 337
+ ++ L N L+G IP + ++ L + L N+ +G IPA G+ NL +V
Sbjct: 730 SKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSNLQLLNVS 909
Query: 338 ENNLTGTIPT 347
NN++G+IPT
Sbjct: 910 FNNISGSIPT 939
Score = 125 bits (315), Expect = 7e-29
Identities = 95/344 (27%), Positives = 159/344 (45%), Gaps = 1/344 (0%)
Frame = +1
Query: 60 NSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFS 119
N + ++ ++L + G++ +FS NL+ L++ N GT+P I + + TL
Sbjct: 16 NIEPLTDLDLSDNFFTGSIPE-SFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLETLLIW 192
Query: 120 LNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPE 179
N GS+P+ + L+ +D S L G IP I L L L N F G
Sbjct: 193 NNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGG--LSS 366
Query: 180 IGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYL 239
I + L L ++ G I + L ++ +DLS N G IP I ++L +
Sbjct: 367 ISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEYFNV 546
Query: 240 AKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPS 299
+ N +L G IP W++ L + +S +P E+ +++ + LD N LSGTIP+
Sbjct: 547 SYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLP-LFESCKSISVIDLDSNSLSGTIPN 723
Query: 300 TIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFE 359
+ + L+ + L N L+G IP + ++ L + N G IP G+ + L +
Sbjct: 724 GVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSNLQLLN 903
Query: 360 VAANKLHGRIPNGLYNITNWFSF-IVSKNDFVGHLPSQICSGGL 402
V+ N + G IP G SF ++ ++ FVG+ S++C L
Sbjct: 904 VSFNNISGSIPTGK-------SFKLMGRSAFVGN--SELCGAPL 1008
Score = 57.8 bits (138), Expect = 2e-08
Identities = 37/122 (30%), Positives = 65/122 (52%), Gaps = 2/122 (1%)
Frame = +1
Query: 59 DNSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNF 118
++ KSIS I+L++ L GT+ + S L+ +N+ NN G IP ++ +I + ++
Sbjct: 658 ESCKSISVIDLDSNSLSGTIPN-GVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDL 834
Query: 119 SLNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLD--LGGNNFVGTPI 176
S N +G IP + + +LQ ++ SF +SG+IP G L+ +G + G P+
Sbjct: 835 SNNKFNGPIPAKFGSSSNLQLLNVSFNNISGSIPT--GKSFKLMGRSAFVGNSELCGAPL 1008
Query: 177 PP 178
P
Sbjct: 1009QP 1014
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 186 bits (473), Expect = 4e-47
Identities = 107/295 (36%), Positives = 171/295 (57%), Gaps = 10/295 (3%)
Frame = +1
Query: 746 KMMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSF 805
K+ F ++++AT F + LIG G G+VYKA+L G VVA+KKL ++ + + F
Sbjct: 43 KLTFADLLDATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQ-----GDREF 207
Query: 806 MSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQA-TAFDWEKRV 864
+E+ET+ I+HRN++ L G+C + LVY++++ GSL +L+ +A +W R
Sbjct: 208 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQKKAGIKLNWAIRR 387
Query: 865 NVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLK--PGLLSWT 922
+ G A L++LHH+C P IIHRD+ S NVLL+ + EA+VSDFG A+ + LS +
Sbjct: 388 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 567
Query: 923 QFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISL-------FLSQST 975
AGT GY PE Q+ + K DVYS+GV+ LE++ GK P D ++ Q
Sbjct: 568 TLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSADFGDNNLVGWVKQHA 747
Query: 976 RLMANNMLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKM 1030
+L +++ +++ + P ++ E++ ++A +CL+ P RPTM QV M
Sbjct: 748 KLKISDIFDPELMKEDPN-----LEMELLQHLKIAVSCLDDRPWRRPTMIQVMAM 897
>TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial (5%)
Length = 1279
Score = 180 bits (456), Expect = 3e-45
Identities = 97/233 (41%), Positives = 141/233 (59%), Gaps = 6/233 (2%)
Frame = +1
Query: 805 FMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRV 864
F E+E L I+HR ++ L G+C+ L+Y +L GGSL + L+ +A DW+ R+
Sbjct: 46 FERELEILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEALHE--RADQLDWDSRL 219
Query: 865 NVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSWTQF 924
N++ G A L+YLHHDCSP IIHRDI S N+LL+ + EA+VSDFG AK L+ T
Sbjct: 220 NIIMGAAKGLAYLHHDCSPRIIHRDIKSSNILLDGNLEARVSDFGLAKLLEDEESHITTI 399
Query: 925 -AGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLMANNML 983
AGTFGY APE Q+ EK DVYSFGVL LE++ GK P D + F+ + ++ L
Sbjct: 400 VAGTFGYLAPEYMQSGRATEKSDVYSFGVLTLEVLSGKRPTD--AAFIEKGPNIV--GWL 567
Query: 984 LIDVLDQRPQHVMKPVDEEVIL-----IARLAFACLNQNPRSRPTMDQVSKML 1031
+ + RP+ ++ P+ E V + + +A C++ +P RPTM +V ++L
Sbjct: 568 NFLITENRPREIVDPLCEGVQMESLDALLSVAIQCVSSSPEDRPTMHRVVQLL 726
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein ,
partial (47%)
Length = 1305
Score = 171 bits (433), Expect = 2e-42
Identities = 105/290 (36%), Positives = 156/290 (53%), Gaps = 7/290 (2%)
Frame = +2
Query: 749 FENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSE 808
F I AT+ F + +G G G VYK L SG VAVK+L +IS + F +E
Sbjct: 50 FSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRL-----SKISGQGGEEFKNE 214
Query: 809 IETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVK 868
+E ++ ++HRN+++L GFC + LVY+F+ SL +L + + DW +R +V+
Sbjct: 215 VEIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVE 394
Query: 869 GVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKF--LKPGLLSWTQFAG 926
G+A + YLH D IIHRD+ + NVLL+ D ++SDFG A+ + + + G
Sbjct: 395 GIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVG 574
Query: 927 TFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGK-----HPGDLISLFLSQSTRLMANN 981
T+GY +PE A E + K DVYSFGVL LEII GK + D+ LS + +L +
Sbjct: 575 TYGYMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLWKDE 754
Query: 982 MLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
L +++DQ + EVI + C+ ++P RPTM V ML
Sbjct: 755 APL-ELMDQSLRE--SYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLML 895
>CO979898
Length = 869
Score = 170 bits (430), Expect = 3e-42
Identities = 90/223 (40%), Positives = 129/223 (57%), Gaps = 1/223 (0%)
Frame = -1
Query: 757 ENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIR 816
+ + K +IG G G VY+ +L +A+K+L+ T E K F E+E ++ I+
Sbjct: 869 QKLNSKDIIGSGGYGVVYELKLDDSTALAIKRLNRGTAER-----DKGFERELEAMADIK 705
Query: 817 HRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSY 876
HRNI+ LHG+ + ++ L+Y+ + GSL L+ ++ DW R + G A +SY
Sbjct: 704 HRNIVTLHGYYTAPLYNLLIYELMPHGSLDSFLHGRSREKVLDWPTRYRIAAGAARGISY 525
Query: 877 LHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLS-WTQFAGTFGYAAPEL 935
LHHDC P IIHRDI S N+LL+ + +A+VSDFG A ++P T AGTFGY APE
Sbjct: 524 LHHDCIPHIIHRDIKSSNILLDQNMDARVSDFGLATLMQPNKTHVSTIVAGTFGYLAPEY 345
Query: 936 AQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLM 978
T K DVYSFGV+ LE++ GK P D F+ + T L+
Sbjct: 344 FDTGRATLKGDVYSFGVVLLELLTGKKPSD--EAFMEEGTMLV 222
>TC226549 UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1627
Score = 168 bits (425), Expect = 1e-41
Identities = 108/302 (35%), Positives = 156/302 (50%), Gaps = 11/302 (3%)
Frame = +3
Query: 747 MMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFM 806
+ + + E T+NF K LIG GS G VY A L++G VAVKKL + ++ E S+ F+
Sbjct: 372 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNNGKAVAVKKLDVSSEPE----SNNEFL 539
Query: 807 SEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDT------QATAFDW 860
+++ +S +++ N ++LHG+C L Y+F GSL +L+ DW
Sbjct: 540 TQVSMVSRLKNGNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDW 719
Query: 861 EKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFL--KPGL 918
+RV + A L YLH PPIIHRDI S NVL+ DY+A+++DF +
Sbjct: 720 IQRVRIAVDAARGLEYLHEKVQPPIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAAR 899
Query: 919 LSWTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLM 978
L T+ GTFGY APE A T ++ +K DVYSFGV+ LE++ G+ P D QS
Sbjct: 900 LHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTW 1079
Query: 979 ANNMLLIDVLDQ--RPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML-AIGK 1035
A L D + Q P+ + + V + +A C+ RP M V K L + K
Sbjct: 1080 ATPRLSEDKVKQCVDPKLKGEYPPKGVAKLGAVAALCVQYEAEFRPNMSIVVKALQPLLK 1259
Query: 1036 SP 1037
SP
Sbjct: 1260 SP 1265
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 166 bits (420), Expect = 5e-41
Identities = 115/366 (31%), Positives = 178/366 (48%), Gaps = 14/366 (3%)
Frame = +3
Query: 680 LDPCGSRKSKNVLRSVLIALGALILVLFGVGISMYTLGRRKKSNEKNQTEEQTQRGVLFS 739
+ P S S + ++++ + +L+ F VGI L RR + ++ +E +
Sbjct: 606 ISPGSSGISAGTIVAIVVPITVAVLI-FIVGICF--LSRRARKKQQGSVKEGK---TAYD 767
Query: 740 IWSHDGKMM-FENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEIS 798
I + D F I AT F +G G G VYK LSSG VVAVK+L + S
Sbjct: 768 IPTVDSLQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRL-----SKSS 932
Query: 799 HFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAF 858
+ F +E+ ++ ++HRN+++L GFC + LVY+++ SL +L +
Sbjct: 933 GQGGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQREL 1112
Query: 859 DWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGL 918
DW +R ++ G+A + YLH D IIHRD+ + N+LL+ D ++SDFG A+
Sbjct: 1113 DWGRRYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQ 1292
Query: 919 L--SWTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGK-----HPGDLISLFL 971
+ ++ GT+GY APE A E + K DVYSFGVL +EI+ GK + D L
Sbjct: 1293 TQGNTSRIVGTYGYMAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLL 1472
Query: 972 SQSTRLMANNMLLIDVLDQRPQHVMKPV------DEEVILIARLAFACLNQNPRSRPTMD 1025
S + +L D P +M P+ EVI + C+ ++P RPTM
Sbjct: 1473 SYAWQLWK---------DGTPLELMDPILRESYNQNEVIRSIHIGLLCVQEDPADRPTMA 1625
Query: 1026 QVSKML 1031
+ ML
Sbjct: 1626 TIVLML 1643
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 165 bits (418), Expect = 9e-41
Identities = 101/285 (35%), Positives = 149/285 (51%), Gaps = 8/285 (2%)
Frame = +2
Query: 755 ATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSG 814
AT+NF +K+++G G G VYK L+ G +VAVK+L EE + F +E+E +S
Sbjct: 143 ATDNFSNKHILGRGGFGKVYKGRLADGSLVAVKRLK----EERTQGG*LQFQTEVEMISM 310
Query: 815 IRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATA-FDWEKRVNVVKGVANA 873
HRN+++L GFC LVY ++ GS+ L ++ W +R + G A
Sbjct: 311 AVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERQESQPPLGWPERKRIALGSARG 490
Query: 874 LSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLK-PGLLSWTQFAGTFGYAA 932
L+YLH C P IIHRD+ + N+LL+ ++EA V DFG AK + T GT G+ A
Sbjct: 491 LAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIA 670
Query: 933 PELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRL------MANNMLLID 986
PE T + +EK DV+ +GV+ LE+I G+ DL L L + + L
Sbjct: 671 PEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKDRKLET 850
Query: 987 VLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
++D Q DEEV + ++A C +P RP M +V +ML
Sbjct: 851 LVDADLQGSYN--DEEVEQLIQVALLCTQGSPMERPKMSEVVRML 979
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,083,418
Number of Sequences: 63676
Number of extensions: 656347
Number of successful extensions: 7947
Number of sequences better than 10.0: 1116
Number of HSP's better than 10.0 without gapping: 4526
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5989
length of query: 1052
length of database: 12,639,632
effective HSP length: 107
effective length of query: 945
effective length of database: 5,826,300
effective search space: 5505853500
effective search space used: 5505853500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC133779.2


