

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.10 + phase: 2 /pseudo
(609 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
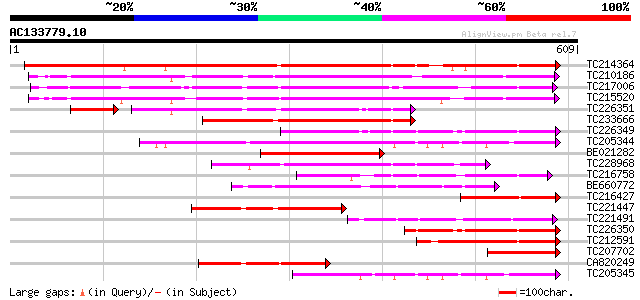
Score E
Sequences producing significant alignments: (bits) Value
TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete 555 e-158
TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete 372 e-103
TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete 368 e-102
TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type proteas... 360 1e-99
TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 212 2e-61
TC233666 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-li... 228 5e-60
TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 223 2e-58
TC205344 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (... 217 1e-56
BE021282 similar to GP|21593457|gb| subtilisin-like serine prote... 194 1e-49
TC228968 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 193 2e-49
TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase... 187 1e-47
BE660772 156 3e-38
TC216427 similar to UP|O82440 (O82440) Subtilisin-like protease ... 150 2e-36
TC221447 similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like p... 149 5e-36
TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 144 1e-34
TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 142 6e-34
TC212591 similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like prot... 142 6e-34
TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like se... 140 2e-33
CA820249 similar to GP|13325079|gb| subtilisin-like protease C1 ... 129 3e-30
TC205345 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (... 129 5e-30
>TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete
Length = 2667
Score = 555 bits (1429), Expect = e-158
Identities = 296/591 (50%), Positives = 384/591 (64%), Gaps = 16/591 (2%)
Frame = +1
Query: 17 FFLFQCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQING 76
F + + Y+VY+GAHSHGP +SVD T SH+D LGS +GS AK++I YSY + ING
Sbjct: 109 FAVKKSYVVYLGAHSHGPELSSVDFNQVTQSHHDFLGSFLGSSNTAKDSIFYSYTRHING 288
Query: 77 FAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDI---NSAWQKGRFGE 133
FAA L+EE A ++AK+PKV+SVF ++ KLHTTRSW+F+ L N + +S W+K RFGE
Sbjct: 289 FAATLDEEVAVEIAKHPKVLSVFENRGRKLHTTRSWDFMELEHNGVIQSSSIWKKARFGE 468
Query: 134 NTIIANIDTGVWPESRSFSDRGIGPIPAKWRG----GSSKKVPCNRKLIGARFFSDAYER 189
II N+DTGVWPES+SFS++G+GPIP+KWRG G CNRKLIGAR+F+ Y
Sbjct: 469 GVIIGNLDTGVWPESKSFSEQGLGPIPSKWRGICHNGIDHTFHCNRKLIGARYFNKGYAS 648
Query: 190 YNGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCW 249
G L +S + RD GHGTHTLSTAGGN V S+F G+GT KGGSP ARVA YKVCW
Sbjct: 649 VAGPLNSSFDSPRDNEGHGTHTLSTAGGNMVARVSVFGQGHGTAKGGSPMARVAAYKVCW 828
Query: 250 SLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNI 309
CF AD+L+A D AI DGVD++SVS GG SST F D V+IG+FHA R +
Sbjct: 829 PPVAGDECFDADILAAFDLAIHDGVDVLSVSLGGSSST----FFKDSVAIGSFHAAKRGV 996
Query: 310 LLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQI-IRGASLFVDLPPNQ 368
++V SAGN GP + N+APW TVAAST+DR F + + +G+ I +G SL ++
Sbjct: 997 VVVCSAGNSGPAEATAENLAPWHVTVAASTMDRQFPTYVVLGNDITFKGESLSATKLAHK 1176
Query: 369 SFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGA 428
+ ++ + DAK ++A DA C+ TLDP+K KGKIV C R G V +G++A AGA
Sbjct: 1177FYPIIKATDAKLASARAEDAVLCQNGTLDPNKAKGKIVVCLR-GINARVDKGEQAFLAGA 1353
Query: 429 KGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATD---TIESGTKIRFS 485
GM L N K +GN ++++PHVL P + TD + +F
Sbjct: 1354VGMVLAND-KTTGNEIIADPHVL--------------PASHINFTDGSAVFKYINSTKFP 1488
Query: 486 QAI-----TLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLL 540
A T + KPAP MA+FSS+GPN + P ILKPD+TAPGV+++AAY+ +N +
Sbjct: 1489VAYITHPKTQLDTKPAPFMAAFSSKGPNTIVPEILKPDITAPGVSVIAAYTEAQGPTNQV 1668
Query: 541 TDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
D RR PFN + GTSMSCPHV+G GL++ L+P WS AAIKSAIMTT T
Sbjct: 1669FDKRR-IPFNSVSGTSMSCPHVSGIVGLLRALYPTWSTAAIKSAIMTTATT 1818
>TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete
Length = 2401
Score = 372 bits (955), Expect = e-103
Identities = 222/580 (38%), Positives = 335/580 (57%), Gaps = 10/580 (1%)
Frame = +1
Query: 21 QCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAM 80
+ YIVYMGA S D + + H +L S++ E A ++ +Y +GFAA
Sbjct: 175 EVYIVYMGAAD------STDA-SFRNDHAQVLNSVLRRNENA---LVRNYKHGFSGFAAR 324
Query: 81 LEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANI 140
L ++EA +A+ P VVSVF KLHTTRSW+FL +++I +
Sbjct: 325 LSKKEATSIAQKPGVVSVFPGPVLKLHTTRSWDFLKYQTQVKIDTKPNAVSKSSSVIGIL 504
Query: 141 DTGVWPESRSFSDRGIGPIPAKWRGGSSKKVP-----CNRKLIGARFFSDAYERYNGKLP 195
DTG+WPE+ SFSD+G+GP+P++W+G K CNRKLIGAR+++D +
Sbjct: 505 DTGIWPEAASFSDKGMGPVPSRWKGTCMKSQDFYSSNCNRKLIGARYYADPND------- 663
Query: 196 TSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAA 255
+ TARD GHGTH TA G V AS + + G KGGSP +R+A Y+VC +
Sbjct: 664 SGDNTARDSNGHGTHVAGTAAGVMVTNASYYGVATGCAKGGSPESRLAVYRVCSNF---- 831
Query: 256 SCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASA 315
C G+ +L+A D AI DGVD++SVS G S+ ++ +D +S+GAFHA+ IL+V SA
Sbjct: 832 GCRGSSILAAFDDAIADGVDLLSVSLGA-STGFRPDLTSDPISLGAFHAMEHGILVVCSA 1008
Query: 316 GNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLFVD-LPPNQSFTLV 373
GN+GP+ ++VN APW+ TVAASTIDR+F S I +GD +II+G ++ + L + + L+
Sbjct: 1009GNDGPSSYTLVNDAPWILTVAASTIDRNFLSNIVLGDNKIIKGKAINLSPLSNSPKYPLI 1188
Query: 374 NSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVAC-AREGKIKSVAEGQEALSAGAKGMF 432
AK ++ + +AR C P +LD +KVKGKIV C + K + + + G G+
Sbjct: 1189YGESAKANSTSLVEARQCHPNSLDGNKVKGKIVVCDDKNDKYSTRKKVATVKAVGGIGLV 1368
Query: 433 LENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTK--IRFSQAITL 490
+ + +++ G+ A + + + GVT I S + ++
Sbjct: 1369----------HITDQNEAIASNYGDFPATVIS-SKDGVTILQYINSTSNPVATILATTSV 1515
Query: 491 IGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFN 550
+ KPAP++ +FSSRGP+ + ILKPD+ APGVNILAA+ + + ++ ++ +
Sbjct: 1516LDYKPAPLVPNFSSRGPSSLSSNILKPDIAAPGVNILAAW--IGNGTEVVPKGKKPSLYK 1689
Query: 551 VMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVI 590
++ GTSM+CPHV+G A +KT +P WS ++IKSAIMT+ I
Sbjct: 1690IISGTSMACPHVSGLASSVKTRNPTWSASSIKSAIMTSAI 1809
>TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete
Length = 2422
Score = 368 bits (945), Expect = e-102
Identities = 232/575 (40%), Positives = 326/575 (56%), Gaps = 9/575 (1%)
Frame = +3
Query: 23 YIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAMLE 82
YIVY G + D +A + + +L + S E K + + + + +GF AML
Sbjct: 120 YIVYTGNSMN-------DEASALTLYSSMLQEVADSNAEPK-LVQHHFKRSFSGFVAMLT 275
Query: 83 EEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLG--LHGNDINSAWQKGRFGENTIIANI 140
EEEA ++A++ +VV+VF +K+ +LHTTRSW+F+G L N + + IIA
Sbjct: 276 EEEADRMARHDRVVAVFPNKKKQLHTTRSWDFIGFPLQAN-------RAPAESDVIIAVF 434
Query: 141 DTGVWPESRSFSDRGIGPIPAKWRGG--SSKKVPCNRKLIGARFFSDAYERYNGKLPTSQ 198
D+G+WPES SF+D+G GP P+KW+G +SK CN K+IGA+ + + + K
Sbjct: 435 DSGIWPESESFNDKGFGPPPSKWKGTCQTSKNFTCNNKIIGAKIYK--VDGFFSK--DDP 602
Query: 199 RTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCF 258
++ RD GHGTH STA GN V AS+ +G GT +GG +AR+A YKVCW C
Sbjct: 603 KSVRDIDGHGTHVASTAAGNPVSTASMLGLGQGTSRGGVTKARIAVYKVCWF----DGCT 770
Query: 259 GADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNE 318
AD+L+A D AI DGVDII+VS GG S N F D ++IGAFHA+ +L V SAGN
Sbjct: 771 DADILAAFDDAIADGVDIITVSLGGFSDEN---YFRDGIAIGAFHAVRNGVLTVTSAGNS 941
Query: 319 GPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQI-IRGASLFVDLPPNQSFTLVNSID 377
GP P S+ N +PW +VAASTIDR F + + +G++I G S+ + + ++ D
Sbjct: 942 GPRPSSLSNFSPWSISVAASTIDRKFVTKVELGNKITYEGTSINTFDLKGELYPIIYGGD 1121
Query: 378 A--KFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLEN 435
A K +R+C +LD VKGKIV C E + K++ AGA G ++
Sbjct: 1122APNKGEGIDGSSSRYCSSGSLDKKLVKGKIVLC--ESRSKALG----PFDAGAVGALIQG 1283
Query: 436 Q--PKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGR 493
Q + + L ++ G + I + T T E+ I
Sbjct: 1284QGFRDLPPSLPLPGSYLALQDGASVYDYINSTRTPIATIFKTDETKDTI----------- 1430
Query: 494 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQ 553
APV+ASFSSRGPN V P ILKPD+ APGV+ILA++S + S++ DNR FN++
Sbjct: 1431--APVVASFSSRGPNIVTPEILKPDLVAPGVSILASWSPASPPSDVEGDNRT-LNFNIIS 1601
Query: 554 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
GTSM+CPHV+G A +K+ HP WSPAAI+SA+MTT
Sbjct: 1602GTSMACPHVSGAAAYVKSFHPTWSPAAIRSALMTT 1706
>TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type protease precursor
{Glycine max;} , complete
Length = 2392
Score = 360 bits (924), Expect = 1e-99
Identities = 231/590 (39%), Positives = 335/590 (56%), Gaps = 20/590 (3%)
Frame = +2
Query: 21 QCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAM 80
+ YIVYMGA T L+ + H +L S++ E A ++ +Y +GFAA
Sbjct: 143 EVYIVYMGAADS----TKASLK---NEHAQILNSVLRRNENA---LVRNYKHGFSGFAAR 292
Query: 81 LEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLH-----GNDINSAWQKGRFGENT 135
L +EEA +A+ P VVSVF KLHTTRSW+FL N+ +
Sbjct: 293 LSKEEANSIAQKPGVVSVFPDPILKLHTTRSWDFLKSQTRVNIDTKPNTLSGSSFSSSDV 472
Query: 136 IIANIDTGVWPESRSFSDRGIGPIPAKWRGG--SSKKVP---CNRKLIGARFFSDAYERY 190
I+ +DTG+WPE+ SFSD+G GP+P++W+G +SK CNRK+IGARF+ + E+
Sbjct: 473 ILGVLDTGIWPEAASFSDKGFGPVPSRWKGTCMTSKDFNSSCCNRKIIGARFYPNPEEK- 649
Query: 191 NGKLPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWS 250
TARDF GHGTH STA G V GAS + + GT +GGSP +R+A YKVC +
Sbjct: 650 ---------TARDFNGHGTHVSSTAVGVPVSGASFYGLAAGTARGGSPESRLAVYKVCGA 802
Query: 251 LTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNIL 310
SC G+ +L+ D AI DGVDI+S+S GG T + ++ TD ++IGAFH++ R IL
Sbjct: 803 F---GSCPGSAILAGFDDAIHDGVDILSLSLGGFGGTKT-DLTTDPIAIGAFHSVQRGIL 970
Query: 311 LVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIG-DQIIRGASL-FVDLPPNQ 368
+V +AGN+G P +V+N APW+ TVAASTIDRD S + +G +Q+++G ++ F L +
Sbjct: 971 VVCAAGNDG-EPFTVLNDAPWILTVAASTIDRDLQSDVVLGNNQVVKGRAINFSPLLNSP 1147
Query: 369 SFTLVNSIDAKFSN-ATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAG 427
+ ++ + A +N + DAR C P +LDP KV GKIV C + I + + +
Sbjct: 1148DYPMIYAESAARANISNITDARQCHPDSLDPKKVIGKIVVCDGKNDIYYSTDEKIVIVKA 1327
Query: 428 AKGMFLENQPKVSGNTLL--SEPHVLSTVGGNGQAAI-----TAPPRLGVTATDTIESGT 480
G+ L + SG+ + V +G A + T+ P + AT
Sbjct: 1328LGGIGLVHITDQSGSVAFYYVDFPVTEVKSKHGDAILQYINSTSHPVGTILAT------- 1486
Query: 481 KIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLL 540
+T+ KPAP + FSSRGP+ + +LKPD+ APGVNILAA+ F + ++ +
Sbjct: 1487-------VTIPDYKPAPRVGYFSSRGPSLITSNVLKPDIAAPGVNILAAW--FGNDTSEV 1639
Query: 541 TDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVI 590
R+ + ++ GTSM+ PHV+G A +K +P WS +AIKSAIMT+ I
Sbjct: 1640PKGRKPSLYRILSGTSMATPHVSGLACSVKRKNPTWSASAIKSAIMTSAI 1789
>TC226351 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(48%)
Length = 1490
Score = 212 bits (539), Expect(2) = 2e-61
Identities = 131/314 (41%), Positives = 182/314 (57%), Gaps = 10/314 (3%)
Frame = +3
Query: 132 GENTIIANIDTG-VWPESRSFSDRGIGPIPAKWRGGSSKKVP-----CNRKLIGARFFSD 185
G + II ++T + ++ + ++ G+GP+P+ W+G CNRKLIGARFFS
Sbjct: 585 GSDVIIGVLNTWRLAGKAGASTNTGLGPVPSTWKGACETGTNLTASNCNRKLIGARFFSK 764
Query: 186 AYERYNGKLPTSQ--RTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVA 243
E G + ++ R+ARD GHGTHT STA G+ V AS+F +GT +G + RARVA
Sbjct: 765 GVEAILGPINETEESRSARDDDGHGTHTASTAAGSVVSDASLFGYASGTARGMATRARVA 944
Query: 244 TYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFH 303
YKVCW CF +D+L+AI++AI D V+++S+S GG S + + D V+IGAF
Sbjct: 945 AYKVCWK----GGCFSSDILAAIERAILDNVNVLSLSLGGGMS----DYYRDSVAIGAFS 1100
Query: 304 ALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQI-IRGASLF- 361
A+ IL+ SAGN GP+P S+ NVAPW+ TV A T+DRDF + + +G+ + G SL+
Sbjct: 1101AMENGILVSCSAGNAGPSPYSLSNVAPWITTVGAGTLDRDFPAYVALGNGLNFSGVSLYR 1280
Query: 362 VDLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQ 421
+ P+ V + SN + C TL P KV GKIV C R G V +G
Sbjct: 1281GNAVPDSPLPFVYA--GNVSNG-AMNGNLCITGTLSPEKVAGKIVLCDR-GLTARVQKGS 1448
Query: 422 EALSAGAKGMFLEN 435
SAGA GM L N
Sbjct: 1449VVKSAGALGMVLSN 1490
Score = 42.7 bits (99), Expect(2) = 2e-61
Identities = 21/52 (40%), Positives = 32/52 (61%)
Frame = +2
Query: 66 IIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGL 117
I+Y+Y+ I+G+A L EEA L +++V ++LHTTR+ FLGL
Sbjct: 392 IMYTYDNAIHGYATRLTAEEARVLETQAGILAVLPETRYELHTTRTPMFLGL 547
>TC233666 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein,
partial (30%)
Length = 678
Score = 228 bits (582), Expect = 5e-60
Identities = 117/229 (51%), Positives = 151/229 (65%), Gaps = 1/229 (0%)
Frame = +1
Query: 208 GTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAID 267
G+HTLST GG FVPGA++F +GNGT +GGSPRARVATYKVCW D CF AD+++A D
Sbjct: 4 GSHTLSTIGGTFVPGANVFGLGNGTAEGGSPRARVATYKVCWPPIDGNECFDADIMAAFD 183
Query: 268 QAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVN 327
AI DGVD++S+S GG N+ + F D +SIGAFHA + I ++ SAGN GPTP +V N
Sbjct: 184 MAIHDGVDVLSLSLGG----NATDYFDDGLSIGAFHANMKGIPVICSAGNYGPTPATVFN 351
Query: 328 VAPWVFTVAASTIDRDFSSTITI-GDQIIRGASLFVDLPPNQSFTLVNSIDAKFSNATTR 386
VAPW+ TV AST+DR F S + + Q GASL +P ++ + L+N+ DAK +N
Sbjct: 352 VAPWILTVGASTLDRQFDSVVELHNGQRFMGASLSKAMPEDKLYPLINAADAKAANKPVE 531
Query: 387 DARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLEN 435
+A C T+DP K +GKI+ C R G V + AL AGA M L N
Sbjct: 532 NATLCMRGTIDPEKARGKILVCLR-GVTARVEKSLVALEAGAAXMILCN 675
>TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(56%)
Length = 1773
Score = 223 bits (568), Expect = 2e-58
Identities = 136/303 (44%), Positives = 180/303 (58%), Gaps = 2/303 (0%)
Frame = +1
Query: 291 EIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITI 350
+ + D V+IGAF A+ + IL+ SAGN GP P S+ NVAPW+ TV A T+DRDF + + +
Sbjct: 1 DYYRDSVAIGAFSAMEKGILVSCSAGNSGPGPYSLSNVAPWITTVGAGTLDRDFPAYVAL 180
Query: 351 GDQI-IRGASLFV-DLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVAC 408
G+ + G SL+ + P+ S LV + SN + C TL P KV GKIV C
Sbjct: 181 GNGLNFSGVSLYRGNALPDSSLPLVYA--GNVSNGAM-NGNLCITGTLSPEKVAGKIVLC 351
Query: 409 AREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRL 468
R G V +G SAGA GM L N +G L+++ H+L +A L
Sbjct: 352 DR-GLTARVQKGSVVKSAGALGMVLSNTA-ANGEELVADAHLLPATAVGQKAGDAIKKYL 525
Query: 469 GVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILA 528
A T+ KI F T +G +P+PV+A+FSSRGPN + P ILKPD+ APGVNILA
Sbjct: 526 VSDAKPTV----KIFFEG--TKVGIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILA 687
Query: 529 AYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
+S + L DNRR FN++ GTSMSCPHV+G A LIK+ HP+WSPAA++SA+MTT
Sbjct: 688 GWSKAVGPTGLPVDNRR-VDFNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTT 864
Query: 589 VIT 591
T
Sbjct: 865 AYT 873
>TC205344 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (AT4g30020
protein) (AT4g30020/F6G3_50), partial (78%)
Length = 2192
Score = 217 bits (552), Expect = 1e-56
Identities = 169/486 (34%), Positives = 246/486 (49%), Gaps = 34/486 (6%)
Frame = +2
Query: 140 IDTGVWPESRSFSDRGI---GPIPAKWRGG-----SSKKVPCNRKLIGARFFSDAYERYN 191
+D+G++P SF+ GP+ +++RG +KK CN K++GA+ F+ A
Sbjct: 8 VDSGIYPHHPSFTTHNTEPYGPV-SRYRGKCEVDPDTKKSFCNGKIVGAQHFAQAAIAAG 184
Query: 192 GKLPTSQ-RTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWS 250
P + D GHG+HT S A G + G G +PRAR+A YK +
Sbjct: 185 AFNPFIVFDSPLDGDGHGSHTASIAAGRNGIPVRMHGHEFGKASGMAPRARIAVYKALYR 364
Query: 251 LTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSS-TNSEEIFTDEVSIGAFHALARNI 309
L F ADV++AIDQA+ DGVDI+S+S G S +N++ F + A+ +
Sbjct: 365 LFGG---FIADVVAAIDQAVHDGVDILSLSVGPNSPPSNTKTTFLNPFDATLLGAVKAGV 535
Query: 310 LLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLFVDLPPNQ 368
+ +AGN GP P S+V+ +PW+ TVAA+ DR + + + +G+ +I+ G L NQ
Sbjct: 536 FVAQAAGNGGPFPKSLVSYSPWIATVAAAIDDRRYKNHLILGNGKILAGLGLSPSTRLNQ 715
Query: 369 SFTLVNSIDAKFSNATTR-DARFC-RPRTLDPSKVKGKIVACARE-------GKIKSVAE 419
++TLV + D ++ T+ C RP L+ + +KG I+ C IK V+E
Sbjct: 716 TYTLVAATDVLLDSSVTKYSPTDCQRPELLNKNLIKGNILLCGYSYNFVIGSASIKQVSE 895
Query: 420 GQEALSAGAKGMFLENQPKVSGNTLLSE-----PHVLSTVGGNGQAAIT----APPRLGV 470
+AL A +++EN VS T P +L T + I + PR
Sbjct: 896 TAKALGAVGFVLYVEN---VSPGTKFDPVPVGIPGILITDASKSKELIDYYNISTPRDWT 1066
Query: 471 TATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQV-----QPYILKPDVTAPGVN 525
T E KI + I K AP +A FS+RGPN + +LKPD+ APG
Sbjct: 1067GRVKTFEGTGKIE--DGLMPILHKSAPQVAMFSARGPNIKDFSFQEADLLKPDILAPGSL 1240
Query: 526 ILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAI 585
I AA+SL + N G F ++ GTSM+ PH+AG A LIK HP+WSPAAIKSA+
Sbjct: 1241IWAAWSLNGTDE----PNYAGEGFAMISGTSMAAPHIAGIAALIKQKHPHWSPAAIKSAL 1408
Query: 586 MTTVIT 591
MTT T
Sbjct: 1409MTTSTT 1426
>BE021282 similar to GP|21593457|gb| subtilisin-like serine protease
{Arabidopsis thaliana}, partial (5%)
Length = 402
Score = 194 bits (493), Expect = 1e-49
Identities = 95/133 (71%), Positives = 113/133 (84%)
Frame = +1
Query: 270 IDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVA 329
IDDGVDIIS+SAGG E IFTDEVSIGAFHA+ARN +LVASAGN+GPTPG+V+NVA
Sbjct: 1 IDDGVDIISLSAGGSYVVTPEGIFTDEVSIGAFHAIARNRILVASAGNDGPTPGTVLNVA 180
Query: 330 PWVFTVAASTIDRDFSSTITIGDQIIRGASLFVDLPPNQSFTLVNSIDAKFSNATTRDAR 389
PWVFT+AAST+DRDFSS +TI ++ I GASLFV+LPPN++F+L+ + DAK +NAT RDA
Sbjct: 181 PWVFTIAASTLDRDFSSNLTINNRQITGASLFVNLPPNKAFSLILATDAKLANATFRDAE 360
Query: 390 FCRPRTLDPSKVK 402
CRP TLDP KVK
Sbjct: 361 LCRPGTLDPEKVK 399
>TC228968 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (20%)
Length = 1008
Score = 193 bits (491), Expect = 2e-49
Identities = 119/306 (38%), Positives = 167/306 (53%), Gaps = 6/306 (1%)
Frame = +3
Query: 217 GNFVPGAS-IFNIGNGTIKGGSPRARVATYKVCWSLTDAAS-----CFGADVLSAIDQAI 270
G VP AS I GT GG+P AR+A YK CW + + C D+L AID AI
Sbjct: 12 GRVVPNASAIGGFAKGTALGGAPLARLAIYKACWPIKGKSKHEGNICTNIDMLKAIDDAI 191
Query: 271 DDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAP 330
DGVD++S+S G + + EE D ++ GA HA+ +NI++V SAGN GP P ++ N AP
Sbjct: 192 GDGVDVLSISIGFSAPISYEE---DVIARGALHAVRKNIVVVCSAGNSGPLPQTLSNPAP 362
Query: 331 WVFTVAASTIDRDFSSTITIGDQIIRGASLFVDLPPNQSFTLVNSIDAKFSNATTRDARF 390
W+ TVAAST+DR F + I + II G S+ N + LV + D + + ++ F
Sbjct: 363 WIITVAASTVDRSFHAPIKLNGTIIEGRSITPLHMGNSFYPLVLARDVEHPGLPSNNSGF 542
Query: 391 CRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHV 450
C TL P+K +GKIV C R G+ + + +G E AG G L N K++G + S+PH
Sbjct: 543 CLDNTLQPNKARGKIVLCMR-GQGERLKKGLEVQRAGGVGFILGNN-KLNGKDVPSDPHF 716
Query: 451 LSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQV 510
+ G + + ++ + T + T++ KPAP MASFSSRGPN V
Sbjct: 717 IPATGVSYENSLKLIQYVHSTPNPMAQ------ILPGTTVLETKPAPSMASFSSRGPNIV 878
Query: 511 QPYILK 516
P ILK
Sbjct: 879 DPNILK 896
>TC216758 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(35%)
Length = 796
Score = 187 bits (475), Expect = 1e-47
Identities = 119/288 (41%), Positives = 161/288 (55%), Gaps = 13/288 (4%)
Frame = +3
Query: 309 ILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLFVDLP-- 365
+ + +SAGN+GP+ SV N+APW+ TV A TIDRDF S + +GD + + G SL+
Sbjct: 6 VFVSSSAGNDGPSGMSVTNLAPWLTTVGAGTIDRDFPSQVILGDGRRLSGVSLYAGAALK 185
Query: 366 --------PNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSV 417
P +S L +S+ C +LDP+ VKGKIV C R G V
Sbjct: 186 GKMYQLVYPGKSGILGDSL--------------CMENSLDPNMVKGKIVICDR-GSSPRV 320
Query: 418 AEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLST--VGGNGQAAITAPPRLGVTATDT 475
A+G AG GM L N +G L+ + H+L VG N I T T
Sbjct: 321 AKGLVVKKAGGVGMILANGIS-NGEGLVGDAHLLPACAVGANEGDVIKKYISSSTNPTAT 497
Query: 476 IESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFAS 535
++ T++G KPAPV+ASFS+RGPN + P ILKPD APGVNILAA++
Sbjct: 498 LDFKG--------TILGIKPAPVIASFSARGPNGLNPQILKPDFIAPGVNILAAWTQAVG 653
Query: 536 ASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKS 583
+ L +D RR FN++ GTSM+CPHV+G A L+K+ HP+WSPAA++S
Sbjct: 654 PTGLDSDTRR-TEFNILSGTSMACPHVSGAAALLKSAHPDWSPAALRS 794
>BE660772
Length = 826
Score = 156 bits (394), Expect = 3e-38
Identities = 104/290 (35%), Positives = 154/290 (52%), Gaps = 2/290 (0%)
Frame = +2
Query: 239 RARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVS 298
+AR+A YK+CW L CF +D+L+A+D+A+ DGV +IS+S G SS + + + D ++
Sbjct: 2 KARIAAYKICWKL----GCFDSDILAAMDEAVSDGVHVISLSVG--SSGYAPQYYRDSIA 163
Query: 299 IGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRG 357
+GAF A N+L+ SAGN GP P + VN+APW+ TV AST+DR+F + + +GD ++ G
Sbjct: 164 VGAFGAAKHNVLVSCSAGNSGPGPSTAVNIAPWILTVGASTVDREFPADVILGDGRVFGG 343
Query: 358 ASLFV-DLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKS 416
SL+ + P+ LV + D +R+C +L+ SKV+GKI C R G +
Sbjct: 344 VSLYYGESLPDFKLPLVYAKDC--------GSRYCYIGSLESSKVQGKIXVCDRGGNAR- 496
Query: 417 VAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTI 476
V +G G GM + N + L + I R + +
Sbjct: 497 VEKGSAVKLTGGLGMIMGNT*SQR*KSFLQMRIF*QLLWWVKPQEIRL--RSXXSLSQVP 670
Query: 477 ESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNI 526
S I + L G P +ASFSSRGPN ++ ILKP+V GVNI
Sbjct: 671 NSNDXI*XNW--ELGGLXLXPXVASFSSRGPNHLRWKILKPEVLVLGVNI 814
>TC216427 similar to UP|O82440 (O82440) Subtilisin-like protease (Fragment),
partial (15%)
Length = 689
Score = 150 bits (378), Expect = 2e-36
Identities = 75/107 (70%), Positives = 85/107 (79%)
Frame = +2
Query: 485 SQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNR 544
S A T IG KPAP++A FSSRGP+ VQP ILKPD+TAPGVN++AA++ A SNL +D R
Sbjct: 146 SAAETYIGVKPAPIIAGFSSRGPSSVQPLILKPDITAPGVNVIAAFTQGAGPSNLPSDRR 325
Query: 545 RGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
R FNV QGTSMSCPHVAG AGL+KT HP WSPAAIKSAIMTT T
Sbjct: 326 RSL-FNVQQGTSMSCPHVAGIAGLLKTYHPTWSPAAIKSAIMTTATT 463
>TC221447 similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like proteinase,
partial (21%)
Length = 491
Score = 149 bits (375), Expect = 5e-36
Identities = 78/167 (46%), Positives = 110/167 (65%), Gaps = 1/167 (0%)
Frame = +1
Query: 196 TSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAA 255
T + RD GHG+HT +TA G+ V GAS+F NGT +G + +AR+ATYKVCW
Sbjct: 1 TESXSPRDDDGHGSHTSTTAAGSAVVGASLFGFANGTARGMATQARLATYKVCW----LG 168
Query: 256 SCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASA 315
CF +D+ + ID+AI+DGV+I+S+S GG + + D ++IG F A A IL+ SA
Sbjct: 169 GCFTSDIAAGIDKAIEDGVNILSMSIGG----GLMDYYKDTIAIGTFAATAHGILVSNSA 336
Query: 316 GNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLF 361
GN GP+ ++ NVAPW+ TV A TIDRDF + IT+G+ ++ G SL+
Sbjct: 337 GNGGPSQATLSNVAPWLTTVGAGTIDRDFPAYITLGNGKMYTGVSLY 477
>TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (34%)
Length = 872
Score = 144 bits (363), Expect = 1e-34
Identities = 90/225 (40%), Positives = 124/225 (55%)
Frame = +3
Query: 364 LPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEA 423
LPPN +V + +N + C TL KV GKIV C R G + V +G
Sbjct: 15 LPPNSPLPIVYA-----ANVSDESQNLCTRGTLIAEKVAGKIVICDRGGNAR-VEKGLVV 176
Query: 424 LSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIR 483
SAG GM L N G L+++ ++L +++ + + T + G
Sbjct: 177 KSAGGIGMILSNNEDY-GEELVADSYLLPAAALGQKSSNELKKYVFSSPNPTAKLGF--- 344
Query: 484 FSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDN 543
T +G +P+PV+A+FSSRGPN + P ILKPD+ APGVNILA ++ + L T++
Sbjct: 345 ---GGTQLGVQPSPVVAAFSSRGPNVLTPKILKPDLIAPGVNILAGWTGAVGPTGL-TED 512
Query: 544 RRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
R FN++ GTSMSCPHV G A L+K HP WSPAAI+SA+MTT
Sbjct: 513 TRHVEFNIISGTSMSCPHVTGLAALLKGTHPEWSPAAIRSALMTT 647
>TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 1312
Score = 142 bits (357), Expect = 6e-34
Identities = 81/167 (48%), Positives = 105/167 (62%)
Frame = +1
Query: 425 SAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRF 484
SAGA GM L N +G L+++ H+L +A L A T+ KI F
Sbjct: 7 SAGALGMVLSNTA-ANGEELVADAHLLPATAVGQKAGDAIKKYLFSDAKPTV----KILF 171
Query: 485 SQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNR 544
T +G +P+PV+A+FSSRGPN + P ILKPD+ APGVNILA +S + L DNR
Sbjct: 172 EG--TKLGIQPSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLPVDNR 345
Query: 545 RGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
R FN++ GTSMSCPHV+G A LIK+ HP+WSPAA++SA+MTT T
Sbjct: 346 R-VDFNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYT 483
>TC212591 similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein, partial
(19%)
Length = 463
Score = 142 bits (357), Expect = 6e-34
Identities = 82/154 (53%), Positives = 96/154 (62%)
Frame = +1
Query: 438 KVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAP 497
++SGN L+++PH+L + I L V A T + KPAP
Sbjct: 19 ELSGNELIADPHLLPA------SQINYKDGLAVYAFMNSTKNPLGYIYPPKTKLQIKPAP 180
Query: 498 VMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSM 557
MA+FSSRGPN V P ILKPDV APGVNI+AAYS S +NL D RR PF M GTSM
Sbjct: 181 AMAAFSSRGPNTVTPEILKPDVIAPGVNIIAAYSEGVSPTNLGFDKRR-VPFITMSGTSM 357
Query: 558 SCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVIT 591
SCPHVAG GL+KTLHP+WSPA IKSA+MTT T
Sbjct: 358 SCPHVAGVVGLLKTLHPDWSPAVIKSALMTTART 459
>TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like serine
protease AIR3 (Fragment), partial (20%)
Length = 1025
Score = 140 bits (352), Expect = 2e-33
Identities = 69/78 (88%), Positives = 71/78 (90%)
Frame = +2
Query: 514 ILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLH 573
ILKPDVTAPGVNILAAYS ASASNLL DNRRGF FNV+QGTS+SCPHVAG AGLIKTLH
Sbjct: 5 ILKPDVTAPGVNILAAYSELASASNLLVDNRRGFKFNVLQGTSVSCPHVAGIAGLIKTLH 184
Query: 574 PNWSPAAIKSAIMTTVIT 591
PNWSPAAIKSAIMTT T
Sbjct: 185 PNWSPAAIKSAIMTTATT 238
>CA820249 similar to GP|13325079|gb| subtilisin-like protease C1 {Glycine
max}, partial (15%)
Length = 421
Score = 129 bits (325), Expect = 3e-30
Identities = 72/143 (50%), Positives = 88/143 (61%), Gaps = 1/143 (0%)
Frame = +2
Query: 203 DFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADV 262
D GHG+H STA GN V AS+ G+GT +GG P AR+A YKVCW A C D+
Sbjct: 5 DTTGHGSHCASTAAGNPVRSASLLGFGSGTARGGVPSARIAVYKVCW----ATGCDTTDI 172
Query: 263 LSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPT- 321
L A D AI DGVDI+SVS G T++ + F D +IGAFHA+ + IL SA N G
Sbjct: 173 LKAYDAAIADGVDILSVSVGATQLTHN-KYFKDVHAIGAFHAMKKGILTSTSADNLGQLG 349
Query: 322 PGSVVNVAPWVFTVAASTIDRDF 344
P S APW+ +VAASTID+ F
Sbjct: 350 PYSTSKFAPWLLSVAASTIDKKF 418
>TC205345 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (AT4g30020
protein) (AT4g30020/F6G3_50), partial (62%)
Length = 1729
Score = 129 bits (323), Expect = 5e-30
Identities = 102/314 (32%), Positives = 155/314 (48%), Gaps = 26/314 (8%)
Frame = +1
Query: 304 ALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLFV 362
A+ + + +AGN GP P ++V+ +PW+ +VAA+ DR + + + +G+ + + G L
Sbjct: 58 AVKAGVFVAQAAGNHGPLPKTLVSYSPWIASVAAAIDDRRYKNHLILGNGKTLAGIGLSP 237
Query: 363 DLPPNQSFTLVNS----IDAKFSNATTRDARFCRPRTLDPSKVKGKIVACARE------- 411
N+++TLV + +D+ + D + RP L+ + +KG I+ C
Sbjct: 238 STHLNETYTLVAANDVLLDSSLMKYSPTDCQ--RPELLNKNLIKGNILLCGYSFNFVVGT 411
Query: 412 GKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSE-----PHVLSTVGGNGQAAIT--- 463
IK V+E +AL A + +EN +S T + P +L N + I
Sbjct: 412 ASIKKVSETAKALGAVGFVLCVEN---ISLGTKFNPVPVGLPGILIIDVSNSKELIDYYN 582
Query: 464 -APPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQV-----QPYILKP 517
PR + E KI + I K AP +A FS+RGPN + +LKP
Sbjct: 583 ITTPRDWTGRVKSFEGKGKI--GDGLMPILHKSAPQVALFSARGPNIKDFSFQEADLLKP 756
Query: 518 DVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWS 577
D+ APG I AA+ + N G F ++ GTSM+ PH+AG A LIK HP+WS
Sbjct: 757 DILAPGSLIWAAWC----PNGTDEPNYVGEAFAMISGTSMAAPHIAGIAALIKQKHPHWS 924
Query: 578 PAAIKSAIMTTVIT 591
PAAIKSA+MTT T
Sbjct: 925 PAAIKSALMTTSTT 966
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,707,614
Number of Sequences: 63676
Number of extensions: 354256
Number of successful extensions: 3152
Number of sequences better than 10.0: 112
Number of HSP's better than 10.0 without gapping: 2840
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2982
length of query: 609
length of database: 12,639,632
effective HSP length: 103
effective length of query: 506
effective length of database: 6,081,004
effective search space: 3076988024
effective search space used: 3076988024
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC133779.10


