

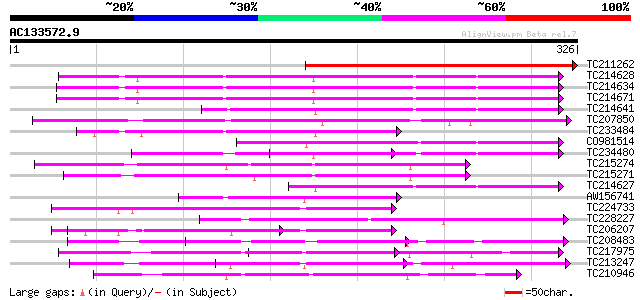
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.9 + phase: 0
(326 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211262 similar to UP|Q9FM86 (Q9FM86) ADP/ATP translocase-like ... 295 2e-80
TC214628 homologue to UP|O49875 (O49875) Adenine nucleotide tran... 211 4e-55
TC214634 homologue to UP|O49447 (O49447) ADP, ATP carrier-like p... 211 4e-55
TC214671 homologue to UP|O49447 (O49447) ADP, ATP carrier-like p... 211 5e-55
TC214641 homologue to UP|O49875 (O49875) Adenine nucleotide tran... 143 1e-34
TC207850 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (63%) 141 5e-34
TC233484 similar to UP|Q8LB08 (Q8LB08) ADP/ATP translocase-like ... 130 6e-31
CO981514 130 1e-30
TC234480 similar to UP|O49875 (O49875) Adenine nucleotide transl... 122 3e-28
TC215274 similar to PIR|T01729|T01729 mitochondrial solute carri... 115 2e-26
TC215271 similar to PIR|T01729|T01729 mitochondrial solute carri... 111 4e-25
TC214627 homologue to UP|O49447 (O49447) ADP, ATP carrier-like p... 108 3e-24
AW156741 similar to GP|14597792|em c_pp004075307r {Physcomitrell... 101 4e-22
TC224733 similar to GB|AAP12888.1|30017309|BT006239 At5g48970 {A... 99 3e-21
TC228227 similar to GB|AAL69531.1|18491125|AY074833 At2g47490/T3... 98 4e-21
TC206207 similar to UP|Q8LCH4 (Q8LCH4) Ca-dependent solute carri... 95 4e-20
TC208483 similar to GB|AAP21144.1|30102452|BT006336 At5g01500/F7... 92 4e-19
TC217975 similar to UP|Q9LY28 (Q9LY28) Peroxisomal Ca-dependent ... 90 2e-18
TC213247 weakly similar to UP|SFC1_YEAST (P33303) Succinate/fuma... 88 5e-18
TC210946 similar to UP|Q9FI43 (Q9FI43) Calcium-binding transport... 87 1e-17
>TC211262 similar to UP|Q9FM86 (Q9FM86) ADP/ATP translocase-like protein,
partial (47%)
Length = 775
Score = 295 bits (754), Expect = 2e-80
Identities = 141/156 (90%), Positives = 151/156 (96%)
Frame = +3
Query: 171 RTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESK 230
R EVRQFRGI+HFLATIF KDGVRGIY+GLPASLHGMV+HRGLYFGGFDT+KE++SEESK
Sbjct: 39 RREVRQFRGIYHFLATIFHKDGVRGIYKGLPASLHGMVVHRGLYFGGFDTMKEIMSEESK 218
Query: 231 PELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGL 290
PELALWKRW+VAQAVTTSAGL+SYPLDTVRRRMMMQSG+E PVYNSTLDCWRKIYRTEGL
Sbjct: 219 PELALWKRWVVAQAVTTSAGLISYPLDTVRRRMMMQSGIEQPVYNSTLDCWRKIYRTEGL 398
Query: 291 ISFYRGAVSNVFRSTGAAAILVLYDEVKKFMNLGRL 326
SFYRGAVSNVFRSTGAAAILVLYDEVKKFMN GR+
Sbjct: 399 ASFYRGAVSNVFRSTGAAAILVLYDEVKKFMNWGRI 506
>TC214628 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
complete
Length = 1561
Score = 211 bits (537), Expect = 4e-55
Identities = 118/295 (40%), Positives = 175/295 (59%), Gaps = 5/295 (1%)
Frame = +3
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIRTV 86
+F D + G V T APIER KLL+Q Q+ ++ +GR +KG+ DC RT+
Sbjct: 345 HFLLDFLMGGVSAAVSKTAAAPIERVKLLIQNQDE---MIKTGRLSEPYKGIGDCFKRTM 515
Query: 87 REEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAA 146
+EG ISLWRGN ++V+RY+P+ ALNF+ KD +K + + D + + N +G A
Sbjct: 516 ADEGAISLWRGNTANVIRYFPTQALNFAFKDYFKRLF-NFKKDRDGYWKWFAGNLASGGA 692
Query: 147 AGCTSLILVYPLDIAHTRLAADIGRTE---VRQFRGIHHFLATIFQKDGVRGIYRGLPAS 203
AG +SL+ VY LD A TRLA D + RQF G+ DGV G+YRG S
Sbjct: 693 AGASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLVDVYRKTLASDGVAGLYRGFNIS 872
Query: 204 LHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRM 263
G++++RGLYFG +D++K ++ S + + + + + +T AGL SYP+DTVRRRM
Sbjct: 873 CVGIIVYRGLYFGLYDSVKPVVLTGSLQD-SFFASFALGWLITNGAGLASYPIDTVRRRM 1049
Query: 264 MMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
MM SG E Y S+LD + +I + EG S ++GA +N+ R+ A +L YD+++
Sbjct: 1050MMTSG-EAVKYKSSLDAFTQILKNEGAKSLFKGAGANILRAVAGAGVLAGYDKLQ 1211
>TC214634 homologue to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (83%)
Length = 1503
Score = 211 bits (537), Expect = 4e-55
Identities = 116/296 (39%), Positives = 177/296 (59%), Gaps = 5/296 (1%)
Frame = +2
Query: 28 NNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIRT 85
+ F D + G V T APIER KLL+Q Q+ ++ SGR +KG+ DC RT
Sbjct: 281 SGFLLDFLMGGVSAAVSKTAAAPIERVKLLIQNQDE---MIKSGRLSEPYKGIGDCFTRT 451
Query: 86 VREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGA 145
+++EGVI+LWRGN ++V+RY+P+ ALNF+ KD +K + + D + + N +G
Sbjct: 452 MKDEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLF-NFKKDKDGYWKWFAGNLASGG 628
Query: 146 AAGCTSLILVYPLDIAHTRLAADIGRTE---VRQFRGIHHFLATIFQKDGVRGIYRGLPA 202
AAG +SL+ VY LD A TRLA D + RQF G+ + DG+ G+YRG
Sbjct: 629 AAGASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLIDVYRKTIKSDGIAGLYRGFNI 808
Query: 203 SLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRR 262
S G++++RGLYFG +D++K ++ + + + +++ +T AGL SYP+DTVRRR
Sbjct: 809 SCVGIIVYRGLYFGMYDSLKPVVLVGGLQD-SFFASFLLGWGITIGAGLASYPIDTVRRR 985
Query: 263 MMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
MMM SG E Y S+L+ ++ I EG S ++GA +N+ R+ A +L YD+++
Sbjct: 986 MMMTSG-EAVKYKSSLEAFKIIVAKEGTKSLFKGAGANILRAVAGAGVLAGYDKLQ 1150
>TC214671 homologue to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (85%)
Length = 1562
Score = 211 bits (536), Expect = 5e-55
Identities = 117/296 (39%), Positives = 176/296 (58%), Gaps = 5/296 (1%)
Frame = +3
Query: 28 NNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGR--RKFKGMFDCIIRT 85
+ F D + G V T APIER KLL+Q Q+ ++ SGR +KG+ DC RT
Sbjct: 303 SGFLVDFLMGGVSAAVSKTAAAPIERVKLLIQNQDE---MIKSGRLSEPYKGIGDCFART 473
Query: 86 VREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGA 145
+++EGVI+LWRGN ++V+RY+P+ ALNF+ KD +K + + D + + N +G
Sbjct: 474 MKDEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLF-NFKKDKDGYWKWFAGNLASGG 650
Query: 146 AAGCTSLILVYPLDIAHTRLAADIGRTE---VRQFRGIHHFLATIFQKDGVRGIYRGLPA 202
AAG +SL+ VY LD A TRLA D + RQF G+ + DGV G+YRG
Sbjct: 651 AAGASSLLFVYSLDYARTRLANDAKAAKKGGERQFNGLVDVYRKTIKSDGVAGLYRGFNI 830
Query: 203 SLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRR 262
S G++++RGLYFG +D++K ++ + + + +++ +T AGL SYP+DTVRRR
Sbjct: 831 SCVGIIVYRGLYFGMYDSLKPVVLVGGLQD-SFFASFLLGWGITIGAGLASYPIDTVRRR 1007
Query: 263 MMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
MMM SG E Y S+L ++ I EG S ++GA +N+ R+ A +L YD+++
Sbjct: 1008MMMTSG-EAVKYKSSLHAFQTIVANEGAKSLFKGAGANILRAVAGAGVLAGYDKLQ 1172
>TC214641 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
partial (57%)
Length = 982
Score = 143 bits (360), Expect = 1e-34
Identities = 81/211 (38%), Positives = 122/211 (57%), Gaps = 3/211 (1%)
Frame = +2
Query: 111 LNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIG 170
LNF+ KD +K + + D + + N +G AAG +SL+ VY LD A TRLA D
Sbjct: 26 LNFAFKDYFKRLFNF-KKDRDGYWKWFAGNLASGGAAGASSLLFVYSLDYARTRLANDAK 202
Query: 171 RTEV---RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSE 227
+ RQF G+ DGV G+YRG S G++++RGLYFG +D++K ++
Sbjct: 203 AAKKGGERQFNGLVDVYRKTLASDGVAGLYRGFNISCVGIIVYRGLYFGLYDSVKPVVLT 382
Query: 228 ESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRT 287
S + + + + + +T AGL SYP+DTVRRRMMM SG E Y S+LD + +I +
Sbjct: 383 GSLQD-SFFASFALGWLITNGAGLASYPIDTVRRRMMMTSG-EAVKYKSSLDAFTQILKN 556
Query: 288 EGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
EG S ++GA +N+ R+ A +L YD+++
Sbjct: 557 EGAKSLFKGAGANILRAVAGAGVLAGYDKLQ 649
>TC207850 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (63%)
Length = 1429
Score = 141 bits (355), Expect = 5e-34
Identities = 101/330 (30%), Positives = 166/330 (49%), Gaps = 20/330 (6%)
Frame = +2
Query: 14 NLKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRR 73
N KRN S + ++L+AG G T VAP+ER K+L QT+ +
Sbjct: 212 NASIKRNESSFDGVPVYVKELIAGGFAGALSKTSVAPLERVKILWQTRTPGFHSL----- 376
Query: 74 KFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNI 133
G++ + + ++ EG + L++GNG+SV+R P AL+F + YKS + +N +
Sbjct: 377 ---GVYQSMNKLLKHEGFLGLYKGNGASVIRIVPYAALHFMTYERYKSWIL---NNYPAL 538
Query: 134 FSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFR-----------GIHH 182
+G + +AG+AAG TS++ YPLD+A T+LA + T + GI
Sbjct: 539 GTGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVADTRGGSIKDGMKGVQPAHNGIKG 718
Query: 183 FLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVA 242
L +++++ GVRG+YRG +L G++ + GL F ++ +K + EE + K M+
Sbjct: 719 VLTSVYKEGGVRGLYRGAGPTLTGILPYAGLKFYMYEKLKTHVPEEHQ------KSIMMR 880
Query: 243 QAVTTSAGL----VSYPLDTVRRRM----MMQSGMEHPVYNSTLDCWRKIYRTEGLISFY 294
+ AGL ++YPLD V+R+M + + E Y +T+D R I +G +
Sbjct: 881 LSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDVRYKNTIDGLRTIVCNQGWKQLF 1060
Query: 295 RGAVSNVFRSTGAAAI-LVLYDEVKKFMNL 323
G N R +AAI YD VK ++ +
Sbjct: 1061HGVSINYIRIVPSAAISFTTYDMVKSWLGI 1150
>TC233484 similar to UP|Q8LB08 (Q8LB08) ADP/ATP translocase-like protein,
partial (63%)
Length = 697
Score = 130 bits (328), Expect = 6e-31
Identities = 76/196 (38%), Positives = 112/196 (56%), Gaps = 9/196 (4%)
Frame = +3
Query: 39 VMGGAVHTI----VAPIERAKLLLQTQESNLAIVASGRRK--FKGMFDCIIRTVREEGVI 92
VMGG I APIER KLLLQ Q ++ G+ K + G+ D R EEG+I
Sbjct: 9 VMGGVAAIISKSAAAPIERVKLLLQNQGE---MIKRGQLKKPYLGVSDGFKRVFMEEGLI 179
Query: 93 SLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSL 152
+ WRG+ ++++RY+P+ A NF+ K +KSI G + D + N +G+AAG T+
Sbjct: 180 AFWRGHQANIIRYFPTQAFNFAFKGYFKSIF-GYSKERDGYIKWFAGNVASGSAAGATTS 356
Query: 153 ILVYPLDIAHTRLAADIGRTEV---RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVI 209
+L+Y LD A TRL D V RQF+G+ DG+ G+YRG S+ G+ +
Sbjct: 357 LLLYHLDYARTRLGTDAIECRVTGQRQFKGLIDVYRKTLSSDGIAGLYRGFGISIWGITL 536
Query: 210 HRGLYFGGFDTIKEML 225
+RG+YFG +DT+K ++
Sbjct: 537 YRGMYFGIYDTMKPIV 584
Score = 40.0 bits (92), Expect = 0.001
Identities = 24/97 (24%), Positives = 47/97 (47%)
Frame = +3
Query: 30 FQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREE 89
F ++ +G+ G ++ ++ A+ L T + +G+R+FKG+ D +T+ +
Sbjct: 309 FAGNVASGSAAGATTSLLLYHLDYARTRLGTDAIECRV--TGQRQFKGLIDVYRKTLSSD 482
Query: 90 GVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGG 126
G+ L+RG G S+ + F + D K I+ G
Sbjct: 483 GIAGLYRGFGISIWGITLYRGMYFGIYDTMKPIVLVG 593
Score = 31.2 bits (69), Expect = 0.66
Identities = 13/40 (32%), Positives = 24/40 (59%)
Frame = +3
Query: 264 MMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFR 303
M++ G Y D +++++ EGLI+F+RG +N+ R
Sbjct: 96 MIKRGQLKKPYLGVSDGFKRVFMEEGLIAFWRGHQANIIR 215
>CO981514
Length = 793
Score = 130 bits (326), Expect = 1e-30
Identities = 77/191 (40%), Positives = 111/191 (57%), Gaps = 3/191 (1%)
Frame = -3
Query: 131 DNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADI--GRTE-VRQFRGIHHFLATI 187
D + + N +GAAAG S + VY LD A TRLA D G+T RQF G+
Sbjct: 758 DGYWKWFAGNMASGAAAGALSSVFVYSLDYARTRLANDAKAGKTGGERQFNGLVDVYRKT 579
Query: 188 FQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTT 247
+ DGV G+YRG S G++++RGLYFG +D++K +L + + L + + VT
Sbjct: 578 LRSDGVAGLYRGFNVSCVGIIVYRGLYFGMYDSLKPVLLVGTLQDSFL-ASFALGWMVTI 402
Query: 248 SAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGA 307
A + SYPLDTVRRRMMM SG E Y S+ D + +I + EG S ++GA +N+ R+
Sbjct: 401 GASIASYPLDTVRRRMMMTSG-EAVKYKSSFDAFSQIVKNEGSKSLFKGAGANILRAVAG 225
Query: 308 AAILVLYDEVK 318
A +L YD+++
Sbjct: 224 AGVLSGYDKLQ 192
>TC234480 similar to UP|O49875 (O49875) Adenine nucleotide translocator,
partial (47%)
Length = 629
Score = 122 bits (305), Expect = 3e-28
Identities = 69/172 (40%), Positives = 100/172 (58%), Gaps = 3/172 (1%)
Frame = +3
Query: 150 TSLILVYPLDIAHTRLAADIGRTE---VRQFRGIHHFLATIFQKDGVRGIYRGLPASLHG 206
TSL VY LD A TRL D + +QF G+ DG+ G+YRG S G
Sbjct: 9 TSLAFVYSLDYARTRLTNDAKGAKNGGQKQFNGLIDCYKKTLASDGIVGLYRGFVISCVG 188
Query: 207 MVIHRGLYFGGFDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQ 266
++++RGLYFG +D+ K +L + LA + + AVT AGL SYP+DT+RRRMMM
Sbjct: 189 IIVYRGLYFGMYDSFKPLLPGTMRDSLA--ANFALGWAVTVVAGLASYPIDTIRRRMMMT 362
Query: 267 SGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
SG + Y S++D ++I EG SF++GA +N+ R+ A +L YD+++
Sbjct: 363 SG-QAVKYKSSIDAGQQILAKEGFRSFFKGAGANILRAVAGAGVLAGYDKLQ 515
Score = 55.1 bits (131), Expect = 4e-08
Identities = 37/152 (24%), Positives = 70/152 (45%)
Frame = +3
Query: 71 GRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNP 130
G+++F G+ DC +T+ +G++ L+RG S + L F + D +K +L P
Sbjct: 87 GQKQFNGLIDCYKKTLASDGIVGLYRGFVISCVGIIVYRGLYFGMYDSFKPLL------P 248
Query: 131 DNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQK 190
+ +ANF G A + + YP+D R+ G + +++ I K
Sbjct: 249 GTMRDSLAANFALGWAVTVVAGLASYPIDTIRRRMMMTSG--QAVKYKSSIDAGQQILAK 422
Query: 191 DGVRGIYRGLPASLHGMVIHRGLYFGGFDTIK 222
+G R ++G A++ V G+ G+D ++
Sbjct: 423 EGFRSFFKGAGANILRAVAGAGV-LAGYDKLQ 515
>TC215274 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (77%)
Length = 1034
Score = 115 bits (289), Expect = 2e-26
Identities = 82/261 (31%), Positives = 130/261 (49%), Gaps = 10/261 (3%)
Frame = +2
Query: 15 LKRKRNSFSSSRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRK 74
L R+ + S F + L+AG V GG T VAP+ER K+LLQ Q + K
Sbjct: 221 LAREGVTAPSYAFTTICKSLVAGGVAGGVSRTAVAPLERLKILLQVQNPHSI-------K 379
Query: 75 FKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNF-SLKDLYKSIL---RGGNSNP 130
+ G + R EG L++GNG++ R P+ A+ F S + K IL R N
Sbjct: 380 YNGTIQGLKYIWRTEGFRGLFKGNGTNCARIVPNSAVKFFSYEQASKGILHLYRKQTGNE 559
Query: 131 DNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQK 190
D + AGA AG ++ YP+D+ R+ ++ Q+RG+ H L+T+ ++
Sbjct: 560 DAQLTPL-LRLGAGACAGIIAMSATYPMDMVRGRITVQTEKSPY-QYRGMFHALSTVLRE 733
Query: 191 DGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEES------KPELALWKRWMVAQA 244
+G R +Y+G S+ G++ + GL F ++++K+ L + + EL++ R A
Sbjct: 734 EGPRALYKGWLPSVIGVIPYVGLNFAVYESLKDWLVKSNPLGLVQDSELSVTTRLACGAA 913
Query: 245 VTTSAGLVSYPLDTVRRRMMM 265
T V+YPLD +RRRM M
Sbjct: 914 AGTIGQTVAYPLDVIRRRMQM 976
Score = 28.9 bits (63), Expect = 3.3
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Frame = +2
Query: 260 RRRMMMQSGMEHPV-YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEV 317
R ++++Q H + YN T+ + I+RTEG ++G +N R +A+ Y++
Sbjct: 335 RLKILLQVQNPHSIKYNGTIQGLKYIWRTEGFRGLFKGNGTNCARIVPNSAVKFFSYEQA 514
Query: 318 KK 319
K
Sbjct: 515 SK 520
>TC215271 similar to PIR|T01729|T01729 mitochondrial solute carrier protein
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (78%)
Length = 1089
Score = 111 bits (278), Expect = 4e-25
Identities = 76/243 (31%), Positives = 122/243 (49%), Gaps = 9/243 (3%)
Frame = +3
Query: 32 RDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGV 91
+ L+AG V GG T VAP+ER K+LLQ Q + K+ G + R EG
Sbjct: 333 KSLVAGGVAGGVSRTAVAPLERLKILLQVQNPH-------NIKYNGTVQGLKYIWRTEGF 491
Query: 92 ISLWRGNGSSVLRYYPSVALNF-SLKDLYKSILRGGNSNPDNIFSGASA--NFVAGAAAG 148
L++GNG++ R P+ A+ F S + K IL N + + AGA AG
Sbjct: 492 RGLFKGNGTNCARIVPNSAVKFFSYEQASKGILHLYKQQTGNEDAQLTPLLRLGAGACAG 671
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
++ YP+D+ R+ + Q+RG+ H L+T+ +++G R +Y+G S+ G++
Sbjct: 672 IIAMSATYPMDMVRGRITVQTEASPY-QYRGMFHALSTVLREEGPRALYKGWLPSVIGVI 848
Query: 209 IHRGLYFGGFDTIKEMLSEES------KPELALWKRWMVAQAVTTSAGLVSYPLDTVRRR 262
+ GL F ++++K+ L + + EL++ R A T V+YPLD +RRR
Sbjct: 849 PYVGLNFAVYESLKDYLIKSNPFDLVENSELSVTTRLACGAAAGTVGQTVAYPLDVIRRR 1028
Query: 263 MMM 265
M M
Sbjct: 1029MQM 1037
Score = 28.1 bits (61), Expect = 5.6
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Frame = +3
Query: 260 RRRMMMQSGMEHPV-YNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEV 317
R ++++Q H + YN T+ + I+RTEG ++G +N R +A+ Y++
Sbjct: 396 RLKILLQVQNPHNIKYNGTVQGLKYIWRTEGFRGLFKGNGTNCARIVPNSAVKFFSYEQA 575
Query: 318 KK 319
K
Sbjct: 576 SK 581
>TC214627 homologue to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (45%)
Length = 912
Score = 108 bits (270), Expect = 3e-24
Identities = 61/161 (37%), Positives = 94/161 (57%), Gaps = 3/161 (1%)
Frame = +2
Query: 161 AHTRLAADIGRTEV---RQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGG 217
A TRLA D + RQF G+ + DGV G+YRG S G++++RGLYFG
Sbjct: 2 ARTRLANDAKAAKKGGERQFNGLVDVYRKTIKSDGVAGLYRGFNISCVGIIVYRGLYFGM 181
Query: 218 FDTIKEMLSEESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNST 277
+D++K ++ + + + +++ +T AGL SYP+DTVRRRMMM SG E Y S+
Sbjct: 182 YDSLKPVVLVGGLQD-SFFASFLLGWGITIGAGLASYPIDTVRRRMMMTSG-EAVKYKSS 355
Query: 278 LDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAILVLYDEVK 318
L ++ I EG S ++GA +N+ R+ A +L YD+++
Sbjct: 356 LHAFQTIVANEGAKSLFKGAGANILRAVAGAGVLAGYDKLQ 478
>AW156741 similar to GP|14597792|em c_pp004075307r {Physcomitrella patens},
partial (34%)
Length = 411
Score = 101 bits (252), Expect = 4e-22
Identities = 53/131 (40%), Positives = 79/131 (59%), Gaps = 3/131 (2%)
Frame = +3
Query: 98 NGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYP 157
N ++V+RY+P+ ALNF+ KD +KS+ G D + + N +G AAG TSL+ VY
Sbjct: 6 NTANVIRYFPTQALNFAFKDYFKSLF--GLKKNDGYWKWFAGNIASGGAAGATSLLFVYS 179
Query: 158 LDIAHTRLAAD---IGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLY 214
LD A TR+A D + RQF G+ DG+ G+YRG S+ G++++RGLY
Sbjct: 180 LDYARTRVANDNKSASKGGSRQFNGLVDVYRKTLASDGIAGLYRGFVPSVVGIIVYRGLY 359
Query: 215 FGGFDTIKEML 225
FG +D+IK ++
Sbjct: 360 FGLYDSIKPVV 392
>TC224733 similar to GB|AAP12888.1|30017309|BT006239 At5g48970 {Arabidopsis
thaliana;} , partial (61%)
Length = 1210
Score = 98.6 bits (244), Expect = 3e-21
Identities = 60/204 (29%), Positives = 101/204 (49%), Gaps = 6/204 (2%)
Frame = +2
Query: 25 SRFNNFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQ---ESNLAIVA---SGRRKFKGM 78
S+ D AGA+ GG T+ +P++ K+ Q Q S+ A++ + K+ GM
Sbjct: 191 SKLKRAMIDSWAGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLRKDLAAASKYTGM 370
Query: 79 FDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGAS 138
F +REEGV WRGN ++L P A+ F++ K+ G + ++I
Sbjct: 371 FQATKDILREEGVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHINLSPY 550
Query: 139 ANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYR 198
++++GA AGC + + YP D+ T LA+ + E + + + I G +G+Y
Sbjct: 551 LSYISGALAGCAATVGSYPFDLLRTILAS---QGEPKVYPNMRSAFMDIVHTRGFQGLYS 721
Query: 199 GLPASLHGMVIHRGLYFGGFDTIK 222
GL +L ++ + GL FG +DT K
Sbjct: 722 GLSPTLVEIIPYAGLQFGTYDTFK 793
>TC228227 similar to GB|AAL69531.1|18491125|AY074833 At2g47490/T30B22.21
{Arabidopsis thaliana;} , partial (61%)
Length = 1098
Score = 98.2 bits (243), Expect = 4e-21
Identities = 67/216 (31%), Positives = 109/216 (50%), Gaps = 4/216 (1%)
Frame = +2
Query: 110 ALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADI 169
A+ FS + KS+L+ +S+ I AN +A + AG + + PL + TRL
Sbjct: 8 AVYFSAYEQLKSLLQSDDSHHLPI----GANMIAASGAGAATTMFTNPLWVVKTRLQTQG 175
Query: 170 GRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEES 229
R V +RG L I ++G+RG+Y GL +L G + H + F ++TIK L+ +
Sbjct: 176 IRPGVVPYRGTLSALRRIAHEEGIRGLYSGLVPALAG-ISHVAIQFPTYETIKFYLANQD 352
Query: 230 KPELALWKRWMVAQAVTTS---AGLVSYPLDTVRRRMMMQSGMEHPVYNSTLDCWRKIYR 286
+ VA A + S A ++YP + VR R+ Q Y+ +DC RK++
Sbjct: 353 DTAMEKLGARDVAIASSVSKIFASTLTYPHEVVRSRLQEQGHHSEKRYSGVIDCIRKVFH 532
Query: 287 TEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFM 321
EG+ FYRG +N+ R+T AA I ++ + +F+
Sbjct: 533 QEGVSGFYRGCATNLLRTTPAAVITFTSFEMIHRFL 640
Score = 37.0 bits (84), Expect = 0.012
Identities = 19/69 (27%), Positives = 35/69 (50%)
Frame = +2
Query: 46 TIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRY 105
T+ P E + LQ Q + +++ G+ DCI + +EGV +RG +++LR
Sbjct: 425 TLTYPHEVVRSRLQEQGHH------SEKRYSGVIDCIRKVFHQEGVSGFYRGCATNLLRT 586
Query: 106 YPSVALNFS 114
P+ + F+
Sbjct: 587 TPAAVITFT 613
>TC206207 similar to UP|Q8LCH4 (Q8LCH4) Ca-dependent solute carrier-like
protein, partial (62%)
Length = 1279
Score = 95.1 bits (235), Expect = 4e-20
Identities = 64/194 (32%), Positives = 103/194 (52%), Gaps = 5/194 (2%)
Frame = +2
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQ--ESNLAIVASGRRKFKGMFDCIIRTVREEGV 91
L+AG V G T AP+ R +L Q Q SN+A + RK +++ R + EEG
Sbjct: 416 LLAGGVAGAFSKTCTAPLARLTILFQIQGMHSNVAAL----RKVS-IWNEASRIIHEEGF 580
Query: 92 ISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGG---NSNPDNIFSGASANFVAGAAAG 148
+ W+GN ++ P ++NF + YK +L+ S+ DN+ + +FV G AG
Sbjct: 581 RAFWKGNLVTIAHRLPYSSVNFYSYEHYKKLLKMVPRLQSHRDNVSADLCVHFVGGGMAG 760
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
T+ YPLD+ TRLAA +T +RGI H L TI +++G+ G+Y+GL +L +
Sbjct: 761 ITAATSTYPLDLVRTRLAA---QTNFTYYRGIWHALHTISKEEGIFGLYKGLGTTLLTVG 931
Query: 209 IHRGLYFGGFDTIK 222
+ F ++T++
Sbjct: 932 PSIAISFSVYETLR 973
Score = 43.1 bits (100), Expect = 2e-04
Identities = 33/140 (23%), Positives = 62/140 (43%), Gaps = 6/140 (4%)
Frame = +2
Query: 25 SRFNNFQRDLMAGAVMGG------AVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGM 78
S +N DL V GG A T + R +L QT + ++G+
Sbjct: 698 SHRDNVSADLCVHFVGGGMAGITAATSTYPLDLVRTRLAAQTNFTY----------YRGI 847
Query: 79 FDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGAS 138
+ + +EEG+ L++G G+++L PS+A++FS+ + +S + S+ + +
Sbjct: 848 WHALHTISKEEGIFGLYKGLGTTLLTVGPSIAISFSVYETLRSYWQSNRSDDSPVVISLA 1027
Query: 139 ANFVAGAAAGCTSLILVYPL 158
++G A+ LV L
Sbjct: 1028CGSLSGIASSTEKESLVQEL 1087
>TC208483 similar to GB|AAP21144.1|30102452|BT006336 At5g01500/F7A7_20
{Arabidopsis thaliana;} , partial (54%)
Length = 675
Score = 91.7 bits (226), Expect = 4e-19
Identities = 68/223 (30%), Positives = 107/223 (47%), Gaps = 3/223 (1%)
Frame = +1
Query: 102 VLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIA 161
V+R P A+ ++YK I +G + + AGA AG TS + YPLD+
Sbjct: 10 VIRVIPYSAVQLFAYEIYKKIFKGKDGELSVL-----GRLAAGAFAGMTSTFITYPLDVL 174
Query: 162 HTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTI 221
RLA + G +R + ++ +++G Y GL SL G+ + + F FD +
Sbjct: 175 RLRLAVEPG------YRTMSEVALSMLREEGFASFYYGLGPSLIGIAPYIAVNFCVFDLL 336
Query: 222 KEMLSE--ESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLD 279
K+ L E + + E +L + A + A L YPLDTVRR+M ++ Y + LD
Sbjct: 337 KKSLPEKYQKRTETSL----LTAVVSASLATLTCYPLDTVRRQMQLRG----TPYKTVLD 492
Query: 280 CWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-LVLYDEVKKFM 321
I +G+I YRG V N ++ ++I L YD VK+ +
Sbjct: 493 AISGIVARDGVIGLYRGFVPNALKNLPNSSIRLTTYDIVKRLI 621
Score = 54.3 bits (129), Expect = 7e-08
Identities = 42/197 (21%), Positives = 82/197 (41%)
Frame = +1
Query: 34 LMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVIS 93
L AGA G I P++ +L L + ++ M + + +REEG S
Sbjct: 112 LAAGAFAGMTSTFITYPLDVLRLRLAVEPG-----------YRTMSEVALSMLREEGFAS 258
Query: 94 LWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLI 153
+ G G S++ P +A+NF + DL K L P+ + + + + +
Sbjct: 259 FYYGLGPSLIGIAPYIAVNFCVFDLLKKSL------PEKYQKRTETSLLTAVVSASLATL 420
Query: 154 LVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGL 213
YPLD ++ + ++ + ++ I +DGV G+YRG + + + +
Sbjct: 421 TCYPLDTVRRQM-----QLRGTPYKTVLDAISGIVARDGVIGLYRGFVPNALKNLPNSSI 585
Query: 214 YFGGFDTIKEMLSEESK 230
+D +K +++ K
Sbjct: 586 RLTTYDIVKRLIAASEK 636
>TC217975 similar to UP|Q9LY28 (Q9LY28) Peroxisomal Ca-dependent solute
carrier-like protein, partial (44%)
Length = 1187
Score = 89.7 bits (221), Expect = 2e-18
Identities = 53/196 (27%), Positives = 99/196 (50%)
Frame = +1
Query: 29 NFQRDLMAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVRE 88
N + +AG + GG T AP++R K++LQ Q +I+ + + + ++
Sbjct: 19 NRSKYFLAGGIAGGISRTATAPLDRLKVVLQVQSEPASIMPA-----------VTKIWKQ 165
Query: 89 EGVISLWRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAG 148
+G++ +RGNG +V++ P A+ F ++ K ++ + N +I G + VAG AG
Sbjct: 166 DGLLGFFRGNGLNVVKVSPESAIKFYAFEMLKKVIGEAHGNKSDI--GTAGRLVAGGTAG 339
Query: 149 CTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMV 208
+ +YP+D+ TRL +E + + I+ ++G R YRGL SL GM+
Sbjct: 340 AIAQAAIYPMDLIKTRLQTC--PSEGGKVPKLGTLTMNIWVQEGPRAFYRGLVPSLLGMI 513
Query: 209 IHRGLYFGGFDTIKEM 224
+ + +DT+K++
Sbjct: 514 PYAAIDLTAYDTMKDI 561
Score = 46.6 bits (109), Expect = 2e-05
Identities = 46/187 (24%), Positives = 80/187 (42%), Gaps = 6/187 (3%)
Frame = +1
Query: 138 SANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQFRGIHHFLATIFQKDGVRGIY 197
S F+AG AG S PLD L + I + I+++DG+ G +
Sbjct: 25 SKYFLAGGIAGGISRTATAPLDRLKVVLQVQ------SEPASIMPAVTKIWKQDGLLGFF 186
Query: 198 RGLPASLHGMVIHRGLYFGGFDTIKEMLSEE--SKPELALWKRWMVAQAVTTSAGLVSYP 255
RG ++ + + F F+ +K+++ E +K ++ R + A YP
Sbjct: 187 RGNGLNVVKVSPESAIKFYAFEMLKKVIGEAHGNKSDIGTAGRLVAGGTAGAIAQAAIYP 366
Query: 256 LDTVRRRMM---MQSGMEHPVYNSTLDCWRKIYRTEGLISFYRGAVSNVFRSTGAAAI-L 311
+D ++ R+ + G + T++ W + EG +FYRG V ++ AAI L
Sbjct: 367 MDLIKTRLQTCPSEGGKVPKLGTLTMNIWVQ----EGPRAFYRGLVPSLLGMIPYAAIDL 534
Query: 312 VLYDEVK 318
YD +K
Sbjct: 535 TAYDTMK 555
Score = 35.0 bits (79), Expect = 0.046
Identities = 20/66 (30%), Positives = 34/66 (51%)
Frame = +3
Query: 58 LQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPSVALNFSLKD 117
LQ Q SN + +KGMFD RT + EG I ++G ++L+ P+ ++ + + +
Sbjct: 678 LQAQPSNTSDA------YKGMFDAFRRTFQLEGFIGFYKGLFPNLLKVVPAASITYVVYE 839
Query: 118 LYKSIL 123
K L
Sbjct: 840 SLKKTL 857
>TC213247 weakly similar to UP|SFC1_YEAST (P33303) Succinate/fumarate
mitochondrial transporter (Regulator of acetyl-CoA
synthetase activity), partial (11%)
Length = 829
Score = 88.2 bits (217), Expect = 5e-18
Identities = 62/213 (29%), Positives = 108/213 (50%), Gaps = 9/213 (4%)
Frame = +3
Query: 119 YKSILRG--GNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRLAADIGRTEVRQ 176
YK++L G + N + +FV G +G TS YPLD+ TRLAA +
Sbjct: 9 YKNVLHSLMGENVSGNSGANLLVHFVGGGLSGITSASATYPLDLVRTRLAA---QRSTMY 179
Query: 177 FRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEMLSEESKPELALW 236
+RGI H +TI + +G G+Y+GL A+L G+ + F ++ ++ + + +
Sbjct: 180 YRGISHAFSTICRDEGFLGLYKGLGATLLGVGPSIAISFAVYEWLRSVWQSQRPDD---- 347
Query: 237 KRWMVAQAVTTSAGLVS----YPLDTVRRRMMMQS-GMEHPVYNSTL-DCWRKIYRTEGL 290
+ +V A + +G+ S +PLD VRRRM ++ G VYN+ L + +I +TEG+
Sbjct: 348 SKAVVGLACGSLSGIASSTATFPLDLVRRRMQLEGVGGRARVYNTGLFGAFGRIIQTEGV 527
Query: 291 ISFYRGAVSNVFRST-GAAAILVLYDEVKKFMN 322
YRG + ++ G + + Y+ +K ++
Sbjct: 528 RGLYRGILPEYYKVVPGVGIVFMTYETLKMLLS 626
Score = 72.4 bits (176), Expect = 3e-13
Identities = 51/197 (25%), Positives = 96/197 (47%), Gaps = 2/197 (1%)
Frame = +3
Query: 35 MAGAVMGGAVHTIVAPIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISL 94
+ G + G + P++ + L Q S + ++G+ R+EG + L
Sbjct: 84 VGGGLSGITSASATYPLDLVRTRLAAQRSTMY--------YRGISHAFSTICRDEGFLGL 239
Query: 95 WRGNGSSVLRYYPSVALNFSLKDLYKSILRGGNSNPDNIFSGASANFVAGAAAGCTSLIL 154
++G G+++L PS+A++F++ + +S+ + + PD+ S A G+ +G S
Sbjct: 240 YKGLGATLLGVGPSIAISFAVYEWLRSVWQ--SQRPDD--SKAVVGLACGSLSGIASSTA 407
Query: 155 VYPLDIAHTRLAAD--IGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRG 212
+PLD+ R+ + GR V G+ I Q +GVRG+YRG+ + +V G
Sbjct: 408 TFPLDLVRRRMQLEGVGGRARVYN-TGLFGAFGRIIQTEGVRGLYRGILPEYYKVVPGVG 584
Query: 213 LYFGGFDTIKEMLSEES 229
+ F ++T+K +LS S
Sbjct: 585 IVFMTYETLKMLLSSIS 635
>TC210946 similar to UP|Q9FI43 (Q9FI43) Calcium-binding transporter-like
protein, partial (42%)
Length = 714
Score = 87.0 bits (214), Expect = 1e-17
Identities = 70/255 (27%), Positives = 115/255 (44%), Gaps = 9/255 (3%)
Frame = +3
Query: 49 APIERAKLLLQTQESNLAIVASGRRKFKGMFDCIIRTVREEGVISLWRGNGSSVLRYYPS 108
AP++R K++LQ Q + I+ + + +K + G++ +RGNG +VL+ P
Sbjct: 21 APLDRLKVVLQIQTTQSHIMPAIKDIWK-----------KGGLLGFFRGNGLNVLKVAPE 167
Query: 109 VALNFSLKDLYKSIL---RGGNSNPDNIFSGASANFVAGAAAGCTSLILVYPLDIAHTRL 165
A+ F ++ K+ + +G + NI GA +AG AG + +YP+D+ TRL
Sbjct: 168 SAIRFYSYEMLKTFITRAKGEEAKAANI--GAMGRLLAGGIAGAVAQTAIYPMDLVKTRL 341
Query: 166 AADIGRTEVRQFRGIHHFLATIFQKDGVRGIYRGLPASLHGMVIHRGLYFGGFDTIKEML 225
++ + + I+ ++G R YRGL SL G++ + G+ ++T+K+M
Sbjct: 342 QTHACKS--GRIPSLGTLSKDIWVQEGPRAFYRGLIPSLLGIIPYAGIDLAAYETLKDMS 515
Query: 226 SE------ESKPELALWKRWMVAQAVTTSAGLVSYPLDTVRRRMMMQSGMEHPVYNSTLD 279
+ E P + L T YPL VR RM Q Y D
Sbjct: 516 KQYILHDGEPGPLVQLG----CGTVSGTLGATCVYPLQVVRTRMQAQRS-----YKGMAD 668
Query: 280 CWRKIYRTEGLISFY 294
+RK EGL FY
Sbjct: 669 VFRKTLEHEGLRGFY 713
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,168,979
Number of Sequences: 63676
Number of extensions: 163668
Number of successful extensions: 990
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 788
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 869
length of query: 326
length of database: 12,639,632
effective HSP length: 98
effective length of query: 228
effective length of database: 6,399,384
effective search space: 1459059552
effective search space used: 1459059552
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC133572.9


