

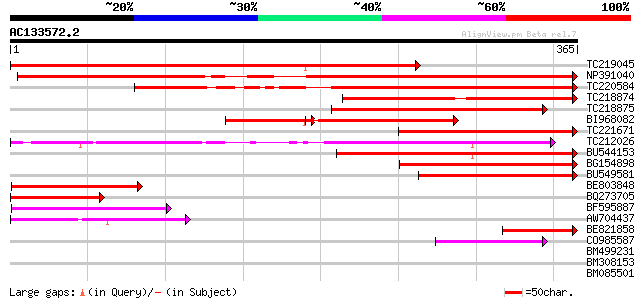
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133572.2 - phase: 0
(365 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219045 similar to UP|IRT3_ARATH (Q8LE59) Fe(II) transport prot... 436 e-122
NP391040 zinc transporter protein ZIP1 [Glycine max] 305 2e-83
TC220584 similar to UP|Q6VM20 (Q6VM20) Metal transport protein, ... 249 1e-66
TC218874 similar to UP|Q6VM17 (Q6VM17) Metal transport protein, ... 244 4e-65
TC218875 similar to UP|Q6VM17 (Q6VM17) Metal transport protein, ... 236 2e-62
BI968082 174 4e-61
TC221671 similar to UP|ZIP4_ARATH (O04089) Zinc transporter 4, c... 211 4e-55
TC212026 similar to UP|Q70II8 (Q70II8) Zinc transporter ZIP6, pa... 196 1e-50
BU544153 similar to PIR|T02478|T02 iron transport protein homolo... 161 4e-40
BG154898 homologue to GP|15418778|gb zinc transporter protein ZI... 125 4e-29
BU549581 homologue to GP|15418778|gb zinc transporter protein ZI... 117 8e-27
BE803848 114 9e-26
BQ273705 similar to GP|14582255|gb| ZIP-like zinc transporter {T... 108 5e-24
BF595887 89 3e-18
AW704437 similar to PIR|T02478|T02 iron transport protein homolo... 64 1e-10
BE821858 similar to GP|15418778|gb| zinc transporter protein ZIP... 54 8e-08
CO985587 53 2e-07
BM499231 similar to GP|13162619|gb zinc transporter {Medicago tr... 39 0.003
BM308153 similar to GP|6960465|gb|A ORF4 hoar {Spodoptera exigua... 36 0.024
BM085501 similar to GP|9759053|dbj AAA-type ATPase-like protein ... 35 0.069
>TC219045 similar to UP|IRT3_ARATH (Q8LE59) Fe(II) transport protein 3,
chloroplast precursor (Iron-regulated transporter 3),
partial (54%)
Length = 1008
Score = 436 bits (1120), Expect = e-122
Identities = 222/276 (80%), Positives = 236/276 (85%), Gaps = 12/276 (4%)
Frame = +1
Query: 1 MTNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVA 60
MTNS+CGGAE +LCRDE AA VLKF+A+ASILL+GMAGIAIPLIGKHRR+LRTDGNLFVA
Sbjct: 181 MTNSSCGGAELELCRDESAALVLKFVAIASILLSGMAGIAIPLIGKHRRFLRTDGNLFVA 360
Query: 61 AKAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGT 120
AKAFAAGVILATGFVHMLSDAT+AL PCLP FPWSKFPFTGFFAM+AAL TLLLDFVGT
Sbjct: 361 AKAFAAGVILATGFVHMLSDATKALRHPCLPAFPWSKFPFTGFFAMLAALLTLLLDFVGT 540
Query: 121 QYYERKQGMNRAVDEQ-ARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHG- 178
QYYERKQGMNRA E+ RVG+SE G GKVFGEEESGGMHI GMHAHAAHHRHNHPHG
Sbjct: 541 QYYERKQGMNRAPSEEPVRVGSSEPGGNGKVFGEEESGGMHIEGMHAHAAHHRHNHPHGN 720
Query: 179 DACDGGGIVKE----------HGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIG 228
DAC G G +KE H H HSH+ I E+TDVRHVVVSQ L+ IVSHSVIIG
Sbjct: 721 DACHGIGNIKEQVHAHSHSHSHSHGHSHSHIEDGEDTDVRHVVVSQXLDFFIVSHSVIIG 900
Query: 229 LSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQ 264
LSLGVSQSPC IRPLIAALSFHQ FEGFALGGCISQ
Sbjct: 901 LSLGVSQSPCTIRPLIAALSFHQLFEGFALGGCISQ 1008
>NP391040 zinc transporter protein ZIP1 [Glycine max]
Length = 1065
Score = 305 bits (781), Expect = 2e-83
Identities = 175/361 (48%), Positives = 221/361 (60%), Gaps = 1/361 (0%)
Frame = +1
Query: 6 CGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAAKAFA 65
C D RD+ A K A+ SIL+AG G+ IPL+GK L + + F KAFA
Sbjct: 88 CTCDREDEERDKSKALRYKIAALVSILVAGAIGVCIPLLGKVISALSPEKDTFFIIKAFA 267
Query: 66 AGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYER 125
AGVIL+TGF+H+L DA E L SPCL E PW +FPFTGF AM A+ TL++D T Y+++
Sbjct: 268 AGVILSTGFIHVLPDAFENLTSPCLKEHPWGEFPFTGFVAMCTAMGTLMVDTYATAYFKK 447
Query: 126 KQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGG 185
+ + DE V E+ESG V +H HA H
Sbjct: 448 H---HHSQDEATDV-------------EKESGHEGHVHLHTHATH--------------- 534
Query: 186 IVKEHGHDHSHALIAANEETDV-RHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLI 244
GH H H ++ +++ RH V+SQVLE+GI+ HS+IIG+SLG S+SP IRPL+
Sbjct: 535 -----GHAHGHVPTDDDQSSELLRHRVISQVLEVGIIVHSIIIGISLGASESPKTIRPLM 699
Query: 245 AALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGA 304
AAL FHQFFEG LG CI+QA FK S T+M FALTTP+G+GIG GI VY+ SP A
Sbjct: 700 AALIFHQFFEGMGLGSCITQANFKKLSITLMGLVFALTTPMGIGIGIGITKVYDENSPTA 879
Query: 305 LIAEGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIW 364
LI EGI +A SAGIL+YMALVDL+AADF++ RM + L+L + L LGAG MS LA W
Sbjct: 880 LIVEGIFNAASAGILIYMALVDLLAADFMNPRMQKSGSLRLGANLSLLLGAGCMSLLAKW 1059
Query: 365 A 365
A
Sbjct: 1060A 1062
>TC220584 similar to UP|Q6VM20 (Q6VM20) Metal transport protein, partial
(71%)
Length = 871
Score = 249 bits (636), Expect = 1e-66
Identities = 139/285 (48%), Positives = 178/285 (61%)
Frame = +3
Query: 81 ATEALNSPCLPEFPWSKFPFTGFFAMMAALFTLLLDFVGTQYYERKQGMNRAVDEQARVG 140
A E+L SPCL E PW KFPFTGF AM++++ TL++D T +Y R+ + +V
Sbjct: 12 AYESLTSPCLKENPWGKFPFTGFVAMLSSIGTLMVDSFATGFYHRQH-----FNPSKQVP 176
Query: 141 TSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHGDACDGGGIVKEHGHDHSHALIA 200
+E G+E +G MH+ H HA H H HG A G +
Sbjct: 177 ADDEE-----MGDEHAGHMHV---HTHATH---GHAHGSAVSPEGSITSEV--------- 296
Query: 201 ANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGG 260
+R ++SQVLE+GIV HSVIIG+SLG + S I+PL+ ALSFHQFFEG LG
Sbjct: 297 ------IRQRIISQVLEIGIVVHSVIIGISLGTAGSIDTIKPLLVALSFHQFFEGMGLGX 458
Query: 261 CISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILV 320
CISQA+F++ ST IMA FF+LTTPIG+ IG G++SVY SP AL EGI ++ SAGIL+
Sbjct: 459 CISQAKFESKSTVIMATFFSLTTPIGIAIGMGVSSVYKENSPTALTVEGIFNSASAGILI 638
Query: 321 YMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
YMALVDL+AADF+S ++ N +LQL L LGAG MS LA WA
Sbjct: 639 YMALVDLLAADFMSPKLQKNLKLQLGQXISLLLGAGCMSLLAKWA 773
>TC218874 similar to UP|Q6VM17 (Q6VM17) Metal transport protein, partial
(39%)
Length = 1029
Score = 244 bits (623), Expect = 4e-65
Identities = 123/151 (81%), Positives = 134/151 (88%)
Frame = +1
Query: 215 VLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTI 274
VLELGIVSHS+IIGLSLGVSQSPC ++ LI ALSFHQFFEGF LGGCISQ QFK S TI
Sbjct: 532 VLELGIVSHSMIIGLSLGVSQSPCTMKALIVALSFHQFFEGFVLGGCISQTQFKTLSATI 711
Query: 275 MACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDLIAADFLS 334
M+CFFALTTP+GV ASV+NPYSPGALI EGILD+LSAGILVYMALVDLIAADFLS
Sbjct: 712 MSCFFALTTPLGV------ASVFNPYSPGALITEGILDSLSAGILVYMALVDLIAADFLS 873
Query: 335 KRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
K+M CNFRLQ++ YC+LFLGAGLMSSLAIWA
Sbjct: 874 KKMPCNFRLQIICYCLLFLGAGLMSSLAIWA 966
>TC218875 similar to UP|Q6VM17 (Q6VM17) Metal transport protein, partial
(37%)
Length = 421
Score = 236 bits (601), Expect = 2e-62
Identities = 116/139 (83%), Positives = 127/139 (90%)
Frame = +2
Query: 208 RHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQF 267
RHVVVSQVLELGIVSHS+IIGLSLGVSQSPC ++PLI ALSFHQFFEGFALGGCISQAQF
Sbjct: 2 RHVVVSQVLELGIVSHSMIIGLSLGVSQSPCTMKPLIVALSFHQFFEGFALGGCISQAQF 181
Query: 268 KASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMALVDL 327
K S TIM+CFFALTTP+GV IG +AS++NPYSP ALI EGILDALSAGILVYMALVDL
Sbjct: 182 KTLSATIMSCFFALTTPLGVAIGASVASIFNPYSPVALITEGILDALSAGILVYMALVDL 361
Query: 328 IAADFLSKRMSCNFRLQLV 346
IAADFLSK+M CNFR Q++
Sbjct: 362 IAADFLSKKMRCNFRFQII 418
>BI968082
Length = 542
Score = 174 bits (442), Expect(2) = 4e-61
Identities = 89/99 (89%), Positives = 91/99 (91%)
Frame = -1
Query: 191 GHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFH 250
GH HSH I E+TDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPC IRPLIAALSFH
Sbjct: 350 GHSHSH--IEDGEDTDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSPCTIRPLIAALSFH 177
Query: 251 QFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGI 289
QFFEGFALGGCISQAQFKASS TIMACFFALTTP+GVGI
Sbjct: 176 QFFEGFALGGCISQAQFKASSATIMACFFALTTPLGVGI 60
Score = 78.6 bits (192), Expect(2) = 4e-61
Identities = 39/63 (61%), Positives = 44/63 (68%), Gaps = 5/63 (7%)
Frame = -2
Query: 140 GTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPHG-DACDGGGIVK----EHGHDH 194
G+SE G VFG+EE+ GMHIVGMHAHAA HRHNH HG DAC G G +K H H H
Sbjct: 541 GSSEPGGNXXVFGKEETVGMHIVGMHAHAAXHRHNHLHGNDACHGIGNIKXXXXXHSHSH 362
Query: 195 SHA 197
SH+
Sbjct: 361 SHS 353
>TC221671 similar to UP|ZIP4_ARATH (O04089) Zinc transporter 4, chloroplast
precursor (ZRT/IRT-like protein 4), partial (30%)
Length = 425
Score = 211 bits (537), Expect = 4e-55
Identities = 104/115 (90%), Positives = 111/115 (96%)
Frame = +2
Query: 251 QFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGI 310
+FFEGFALGGCISQAQFKASS TIMACFFALTTP+GVGIG I+S YNPYSPGALIAEGI
Sbjct: 23 EFFEGFALGGCISQAQFKASSATIMACFFALTTPLGVGIGMAISSGYNPYSPGALIAEGI 202
Query: 311 LDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
LD+LS+GILVYMALVDLIAADFLSKRMSCNFRLQ++SYCMLFLGAGLMSSLAIWA
Sbjct: 203 LDSLSSGILVYMALVDLIAADFLSKRMSCNFRLQILSYCMLFLGAGLMSSLAIWA 367
>TC212026 similar to UP|Q70II8 (Q70II8) Zinc transporter ZIP6, partial (75%)
Length = 1004
Score = 196 bits (499), Expect = 1e-50
Identities = 138/357 (38%), Positives = 191/357 (52%), Gaps = 6/357 (1%)
Frame = +1
Query: 1 MTNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLI--GKHRRYLRTDGNLF 58
+TN+T A CRD A LK I++ I + +AG++ P++ G R D
Sbjct: 97 VTNATRAAA----CRDGAATAHLKMISIFVIFVTSVAGMSSPVVLAGIFRGKPLYD-KAI 261
Query: 59 VAAKAFAAGVILATGFVHMLSDATEAL-NSPCLPEFPWSKFPFTGFFAMMAALFTLLLDF 117
V K FAAGVIL+T VH+L DA AL + PW FPF G ++ AL L++D
Sbjct: 262 VVIKCFAAGVILSTSLVHVLPDAYAALADCHVASRHPWRDFPFAGLVTLVGALLALVVDL 441
Query: 118 VGTQYYERKQGMNRAVDEQARVGTSEEGNVGKVFGEEESGGMHIVGMHAHAAHHRHNHPH 177
+ + E Q + V+++A V E GG
Sbjct: 442 AASSHVE--QHAHAPVEKEAAV---------------ELGGA------------------ 516
Query: 178 GDACDGGGIVKEHGHDHSHALIAANEETDVRHVVVSQVLELGIVSHSVIIGLSLGVSQSP 237
A DG G E G E ++ +VSQVLE+GI+ HSVIIG+++G+SQ+
Sbjct: 517 --AGDGDG---EKGE----------ELAKLKQRLVSQVLEIGIIFHSVIIGVTMGMSQNV 651
Query: 238 CAIRPLIAALSFHQFFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIGTGIASV- 296
C IRPL+AAL+FHQ FEG LGGC++QA F + T M FA+TTPIG+ +G + S+
Sbjct: 652 CTIRPLVAALAFHQIFEGMGLGGCVAQAGFSFGTITFMCFMFAVTTPIGIILGMALFSLT 831
Query: 297 -YNPYSPGALIAEGILDALSAGILVYMALVDLIAADFL-SKRMSCNFRLQLVSYCML 351
Y+ SP ALI EG+L ++S+GIL+YMALVDLIA DF +K M+ L+ S+ L
Sbjct: 832 GYDDSSPNALIMEGLLGSISSGILIYMALVDLIAVDFFHNKLMNSXXLLKKASFVAL 1002
>BU544153 similar to PIR|T02478|T02 iron transport protein homolog F23F1.1 -
Arabidopsis thaliana, partial (45%)
Length = 597
Score = 161 bits (408), Expect = 4e-40
Identities = 85/158 (53%), Positives = 115/158 (71%), Gaps = 3/158 (1%)
Frame = -3
Query: 211 VVSQVLELGIVSHSVIIGLSLGVSQSPCAIRPLIAALSFHQFFEGFALGGCISQAQFKAS 270
+VSQVLE+GI+ HSVIIG+++G+SQ+ C IRP ALSFHQ FEG LGGCI+QA F
Sbjct: 499 LVSQVLEIGIIFHSVIIGVTMGMSQNVCTIRPXXVALSFHQIFEGLGLGGCIAQAGFSFG 320
Query: 271 STTIMACFFALTTPIGVGIGTGIASV--YNPYSPGALIAEGILDALSAGILVYMALVDLI 328
+T M F++TTP+G+ +G + S+ Y+ +P ALI EG+L ++S+GIL+YMALVDLI
Sbjct: 319 TTAYMCFMFSVTTPMGIILGMVLFSMTGYDDTNPKALIMEGLLGSVSSGILIYMALVDLI 140
Query: 329 AADFL-SKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
A DF +K M+ N L+ VS+ L LG+ MS LA+WA
Sbjct: 139 AVDFFHNKLMNSNLYLKKVSFIALTLGSASMSVLALWA 26
>BG154898 homologue to GP|15418778|gb zinc transporter protein ZIP1 {Glycine
max}, partial (31%)
Length = 448
Score = 125 bits (313), Expect = 4e-29
Identities = 68/115 (59%), Positives = 81/115 (70%), Gaps = 1/115 (0%)
Frame = +2
Query: 252 FFEGFALGGCISQAQFKASSTTIMACFFALTTPIGVGIG-TGIASVYNPYSPGALIAEGI 310
FFEG LG CI QA F+ S TIM FFALTTP+G+G GI +VY+ SP ALI EGI
Sbjct: 5 FFEGMGLGSCIIQANFQRLSITIMGLFFALTTPVGIGXWHXGITNVYDENSPTALIVEGI 184
Query: 311 LDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
+A SAGIL+YMALVDL+AADF++ RM + L+L + L LGAG MS LA WA
Sbjct: 185 FNAASAGILIYMALVDLLAADFMNPRMQKSGSLRLGANLSLLLGAGCMSLLAKWA 349
>BU549581 homologue to GP|15418778|gb zinc transporter protein ZIP1 {Glycine
max}, partial (28%)
Length = 594
Score = 117 bits (293), Expect = 8e-27
Identities = 62/102 (60%), Positives = 75/102 (72%)
Frame = -2
Query: 264 QAQFKASSTTIMACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGILDALSAGILVYMA 323
QA F+ S TIM FFALTTP+G+GIG GI +VY+ SP ALI EGI +A SAGIL+YMA
Sbjct: 386 QANFQRLSITIMGLFFALTTPVGIGIGIGITNVYDENSPTALIVEGIFNAASAGILIYMA 207
Query: 324 LVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
LVDL+AADF++ RM + L+L + L LGAG MS LA WA
Sbjct: 206 LVDLLAADFMNPRMQKSGSLRLGANLSLLLGAGCMSLLAKWA 81
>BE803848
Length = 400
Score = 114 bits (284), Expect = 9e-26
Identities = 55/84 (65%), Positives = 70/84 (82%)
Frame = +1
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T+S+C ES+ CRDE AA VLKF+A+ASIL+AG G++IPL+ K RR+LR DG++F AA
Sbjct: 148 TSSSCDITESEQCRDESAAMVLKFVAVASILVAGFGGVSIPLVWKSRRFLRPDGDVFAAA 327
Query: 62 KAFAAGVILATGFVHMLSDATEAL 85
KAFAAGVI+ATGFVHML D+ +AL
Sbjct: 328 KAFAAGVIIATGFVHMLRDSWDAL 399
>BQ273705 similar to GP|14582255|gb| ZIP-like zinc transporter {Thlaspi
caerulescens}, partial (12%)
Length = 420
Score = 108 bits (269), Expect = 5e-24
Identities = 52/61 (85%), Positives = 58/61 (94%)
Frame = +2
Query: 1 MTNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVA 60
MTNS+CGGAE +LCRDE AA VLKF+A+ASILL+GMAGIAIPLIGKHRR+LRTDGNLFVA
Sbjct: 236 MTNSSCGGAELELCRDESAALVLKFVAIASILLSGMAGIAIPLIGKHRRFLRTDGNLFVA 415
Query: 61 A 61
A
Sbjct: 416 A 418
>BF595887
Length = 313
Score = 89.0 bits (219), Expect = 3e-18
Identities = 54/103 (52%), Positives = 61/103 (58%)
Frame = +2
Query: 2 TNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVAA 61
T STC G DLCR+ A LK IA+A I+L MA I++PL+ R LRTD LF A
Sbjct: 2 TTSTCLGPALDLCRN*SNASCLKHIAIAFIILYCMADISLPLML*LRHSLRTDRTLFDTA 181
Query: 62 KAFAAGVILATGFVHMLSDATEALNSPCLPEFPWSKFPFTGFF 104
ILATG HML A+ AL P LP PWSKFPFTG F
Sbjct: 182 NDLYVCFILATGCSHMLFYASNALIHPYLPAIPWSKFPFTGDF 310
>AW704437 similar to PIR|T02478|T02 iron transport protein homolog F23F1.1 -
Arabidopsis thaliana, partial (31%)
Length = 413
Score = 63.5 bits (153), Expect = 1e-10
Identities = 42/120 (35%), Positives = 59/120 (49%), Gaps = 4/120 (3%)
Frame = +3
Query: 1 MTNSTCGGAESDLCRDEPAAFVLKFIAMASILLAGMAGIAIPLIGKHRRYLRTDGNLFVA 60
MT A++ CR+ A LK I++ I + + G+ P+ RY + +A
Sbjct: 57 MTECMTDLAQAAACRNGTTAAHLKVISIFMIFVTSVTGVCAPVT--LARYFQGKSLYNIA 230
Query: 61 A---KAFAAGVILATGFVHMLSDATEALNS-PCLPEFPWSKFPFTGFFAMMAALFTLLLD 116
K FAAGVILAT VH+L DA AL+ + PW FPF G ++ L LL+D
Sbjct: 231 ILLIKCFAAGVILATSLVHVLPDAFAALSDCQVASQHPWKDFPFAGLVTLIGVLMALLVD 410
>BE821858 similar to GP|15418778|gb| zinc transporter protein ZIP1 {Glycine
max}, partial (13%)
Length = 379
Score = 54.3 bits (129), Expect = 8e-08
Identities = 28/48 (58%), Positives = 34/48 (70%)
Frame = -1
Query: 318 ILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIWA 365
IL+YMALVDL AADF+S ++ N +LQL + LGAG MS LA WA
Sbjct: 379 ILIYMALVDLXAADFMSPKLQKNLKLQLGANISXXLGAGCMSLLAKWA 236
>CO985587
Length = 788
Score = 52.8 bits (125), Expect = 2e-07
Identities = 29/73 (39%), Positives = 42/73 (56%), Gaps = 1/73 (1%)
Frame = -3
Query: 275 MACFFALTTPIGVGIGTGIASVYNPYSPGALIAEGIL-DALSAGILVYMALVDLIAADFL 333
M FF L PIG+ IG GI++ +G + SAGIL+Y AL DL+A DF+
Sbjct: 444 MVLFFCLIFPIGIDIGIGISTTTTKAVQRNY*LKGFCYQSASAGILIYTALFDLLATDFM 265
Query: 334 SKRMSCNFRLQLV 346
+ +M +FRL+ +
Sbjct: 264 NTKMLTSFRLKFI 226
>BM499231 similar to GP|13162619|gb zinc transporter {Medicago truncatula},
partial (38%)
Length = 473
Score = 39.3 bits (90), Expect = 0.003
Identities = 30/117 (25%), Positives = 55/117 (46%), Gaps = 2/117 (1%)
Frame = +1
Query: 250 HQFFEGFALGGCISQAQFKASSTTIMA--CFFALTTPIGVGIGTGIASVYNPYSPGALIA 307
H+ F A+G + + K T A FA+++PIGVGIG I + + + A
Sbjct: 43 HKIFAAIAMGIALLRMLPKRPFVTTAAYSLAFAVSSPIGVGIGIAIDATTQGSTADWMFA 222
Query: 308 EGILDALSAGILVYMALVDLIAADFLSKRMSCNFRLQLVSYCMLFLGAGLMSSLAIW 364
I ++ G+ +Y+A+ LI+ F +R + + L + + G ++ + IW
Sbjct: 223 --ITMGIACGVFIYVAINHLISKGFKPQR-TMRYDTPLFRFVAVLSGVAGIAVVMIW 384
>BM308153 similar to GP|6960465|gb|A ORF4 hoar {Spodoptera exigua
nucleopolyhedrovirus}, partial (2%)
Length = 432
Score = 36.2 bits (82), Expect = 0.024
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Frame = +3
Query: 170 HHRHNHPHGDACDGGGIVKE--HGHDHSHAL 198
HH H+H HGD D + K+ HGH H+H +
Sbjct: 288 HHHHHHHHGDDDDAVFVTKQTHHGHAHAHGM 380
>BM085501 similar to GP|9759053|dbj AAA-type ATPase-like protein {Arabidopsis
thaliana}, partial (21%)
Length = 421
Score = 34.7 bits (78), Expect = 0.069
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = +2
Query: 170 HHRHNHPHGDACDGGGIVKEHGHDHSHALI 199
HH H+ P G A G +++GH HSH L+
Sbjct: 95 HHTHHQPQGKARPGTVAARKNGHAHSHVLL 184
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,310,490
Number of Sequences: 63676
Number of extensions: 242026
Number of successful extensions: 2989
Number of sequences better than 10.0: 142
Number of HSP's better than 10.0 without gapping: 2486
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2755
length of query: 365
length of database: 12,639,632
effective HSP length: 99
effective length of query: 266
effective length of database: 6,335,708
effective search space: 1685298328
effective search space used: 1685298328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC133572.2


