

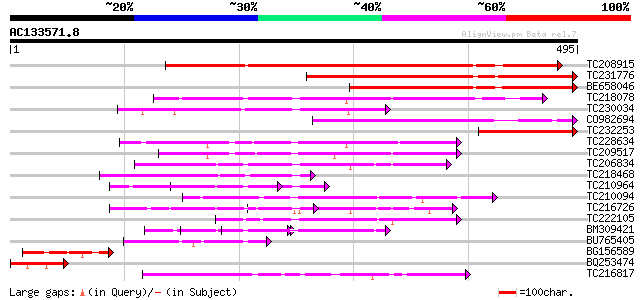
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133571.8 - phase: 0
(495 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208915 489 e-138
TC231776 321 4e-88
BE658046 277 9e-75
TC218078 similar to UP|Q71VM2 (Q71VM2) Plakoglobin/armadillo/bet... 141 8e-34
TC230034 121 6e-28
CO982694 120 2e-27
TC232253 115 6e-26
TC228634 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g6... 109 3e-24
TC209517 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g6... 106 3e-23
TC206834 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, parti... 102 3e-22
TC218468 83 2e-16
TC210964 similar to UP|Q84TU4 (Q84TU4) Arm repeat-containing pro... 79 6e-15
TC210094 weakly similar to UP|Q6K5K9 (Q6K5K9) Avr9/Cf-9 rapidly ... 72 4e-13
TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing pro... 70 2e-12
TC222105 68 8e-12
BM309421 similar to GP|28974687|gb arm repeat-containing protein... 61 1e-09
BU765405 61 1e-09
BG156589 similar to GP|12483839|gb| p6.9 {Heliocoverpa armigera ... 57 1e-08
BQ253474 56 4e-08
TC216817 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like pro... 55 9e-08
>TC208915
Length = 1026
Score = 489 bits (1258), Expect = e-138
Identities = 273/348 (78%), Positives = 299/348 (85%), Gaps = 2/348 (0%)
Frame = +2
Query: 137 KEDSEARGSLAMLGAISPLVGMLD-SEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVH 195
KEDSEAR +LA+LGAI PLVGMLD SED HSQI SLYALLNLGI NDANKAAIVKIGAVH
Sbjct: 5 KEDSEARANLAVLGAIPPLVGMLDDSEDAHSQIASLYALLNLGIGNDANKAAIVKIGAVH 184
Query: 196 KMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSS-KS 254
KMLKLIES DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVR LKNL+ S +S
Sbjct: 185 KMLKLIESSGS-DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRTLKNLNESKIES 361
Query: 255 SSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEG 314
SQ+KQDA+RALYNLSI Q+N+S VLETDLV+FL+++I DMEVSER L+ILSNLVS+PEG
Sbjct: 362 KSQMKQDAMRALYNLSICQSNVSVVLETDLVLFLVSTIGDMEVSERSLAILSNLVSTPEG 541
Query: 315 RKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLEL 374
RKAIS+V DAI +LVD L+WTDSPECQEKASY+LMIMAHKAY DR+ MIEAGIVSSLLEL
Sbjct: 542 RKAISSVSDAIPILVDALSWTDSPECQEKASYVLMIMAHKAYGDRRVMIEAGIVSSLLEL 721
Query: 375 TLVGTALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKTDGGGKE 434
TLVGT LAQKRASRIL+C R+DKGKQVS S GGN L VSAPIC SSSS K
Sbjct: 722 TLVGTTLAQKRASRILECLRIDKGKQVSGSY-GGNFNLGVSAPICGSSSS------SSKG 880
Query: 435 CLMEEVNMMSDEKKAVKQLVQQSLQNNMMKIVKRANLRQDFVPSERFA 482
CL+EE +MS+EKKAVKQLVQQSLQ+NM+KIVKRANLR D V S FA
Sbjct: 881 CLVEEDGIMSEEKKAVKQLVQQSLQSNMIKIVKRANLRHDVVTSTHFA 1024
>TC231776
Length = 832
Score = 321 bits (823), Expect = 4e-88
Identities = 175/237 (73%), Positives = 196/237 (81%), Gaps = 1/237 (0%)
Frame = +3
Query: 260 QDALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAIS 319
QDALRALYNLSI N+SF+LETDLVVFL+NSI DMEV+ER L+ LSN+VS+ EGRKAIS
Sbjct: 3 QDALRALYNLSIFPGNVSFILETDLVVFLVNSIGDMEVTERSLATLSNIVSTREGRKAIS 182
Query: 320 AVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGT 379
V D+I +LVDVLNWTDSPECQEKASYILM+MAHK+Y D+QAMIEAGI SSLLEL+L+G+
Sbjct: 183 TVPDSIPILVDVLNWTDSPECQEKASYILMVMAHKSYGDKQAMIEAGIASSLLELSLLGS 362
Query: 380 ALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKTD-GGGKECLME 438
LAQKRASRIL+ R+DKGKQVS S LG VSAPIC S S K D GGG+EC E
Sbjct: 363 TLAQKRASRILEILRVDKGKQVSGSY---GLGAAVSAPICGSLSG--KPDGGGGRECFEE 527
Query: 439 EVNMMSDEKKAVKQLVQQSLQNNMMKIVKRANLRQDFVPSERFASLTSSSTSKSLPF 495
+ MMS+EKKAVKQLVQQSLQNNM KIVKRANL D VPS+ F SLT SSTSKSLPF
Sbjct: 528 DEEMMSEEKKAVKQLVQQSLQNNMRKIVKRANLPHDIVPSDHFKSLTLSSTSKSLPF 698
>BE658046
Length = 662
Score = 277 bits (708), Expect = 9e-75
Identities = 150/199 (75%), Positives = 167/199 (83%)
Frame = -1
Query: 297 VSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAY 356
VS R L+I SN VS+PEGRKAIS+V DAI +LV+ L+ TDSPECQEKASY+LMIMAHKAY
Sbjct: 662 VSXRXLAIXSNXVSTPEGRKAISSVSDAIPILVNALSXTDSPECQEKASYVLMIMAHKAY 483
Query: 357 ADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSA 416
DR+ MIEAG+VSSLLELTLVGT LAQKRASRIL+C R+DKGKQVS S GGN L VSA
Sbjct: 482 GDRRVMIEAGVVSSLLELTLVGTTLAQKRASRILECLRVDKGKQVSGSY-GGNFNLGVSA 306
Query: 417 PICASSSSLVKTDGGGKECLMEEVNMMSDEKKAVKQLVQQSLQNNMMKIVKRANLRQDFV 476
PIC SSSS GGK CL+ + MMS+EKK VKQLVQQSLQ+NMMKIVKRANLRQDF+
Sbjct: 305 PICGSSSS-----NGGKGCLVVDDGMMSEEKKTVKQLVQQSLQSNMMKIVKRANLRQDFL 141
Query: 477 PSERFASLTSSSTSKSLPF 495
PS FASLTS+STSKSLPF
Sbjct: 140 PSGNFASLTSTSTSKSLPF 84
>TC218078 similar to UP|Q71VM2 (Q71VM2)
Plakoglobin/armadillo/beta-catenin-like protein
(Fragment), partial (88%)
Length = 1310
Score = 141 bits (355), Expect = 8e-34
Identities = 105/350 (30%), Positives = 183/350 (52%), Gaps = 6/350 (1%)
Frame = +3
Query: 126 RIAAAR-VRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDAN 184
R+ AAR +R LTK R L+ A+ PLV ML + S +L ALLNL + ++ N
Sbjct: 210 RLQAARDIRRLTKTSQRCRRQLSQ--AVGPLVSMLRVDSPESHEPALLALLNLAVKDEKN 383
Query: 185 KAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRI 244
K IV+ GA+ ++ ++S + ++ E+ A+ L LSA +NKPII + G IP LV+I
Sbjct: 384 KINIVEAGALEPIISFLKSQ---NLNLQESATASLLTLSASSTNKPIISACGVIPLLVQI 554
Query: 245 LKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSI----EDMEVSER 300
L++ S Q K DA+ AL NLS + N+S +LET+ + ++++ + + + +E+
Sbjct: 555 LRD------GSHQAKADAVMALSNLSTHTNNLSIILETNPIPYMVDLLKTCKKSSKTAEK 716
Query: 301 VLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYAD-R 359
+++ +LV EGR A+++ + + +V+VL + + + +E A L+ M R
Sbjct: 717 CCALIESLVDYDEGRTALTSEEGGVLAVVEVLE-SGTLQSREHAVGALLTMCQSDRCKYR 893
Query: 360 QAMIEAGIVSSLLELTLVGTALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPIC 419
+ ++ G++ LLELT+ GT +Q +A +LQ R + D T+ +C
Sbjct: 894 EPILREGVIPGLLELTVQGTPKSQSKARTLLQLLRESPYPRSEIQPD------TLENIVC 1055
Query: 420 ASSSSLVKTDGGGKECLMEEVNMMSDEKKAVKQLVQQSLQNNMMKIVKRA 469
S + D GK KK + ++VQ S++ ++ + +RA
Sbjct: 1056NIISQIDGDDQSGK------------AKKMLAEMVQVSMEQSLRHLQQRA 1169
>TC230034
Length = 916
Score = 121 bits (304), Expect = 6e-28
Identities = 90/251 (35%), Positives = 135/251 (52%), Gaps = 13/251 (5%)
Frame = +1
Query: 95 ETNAESKKKEETLTEMKHVV------KDLRGEDSTKRRIAAARVRSLTKEDSEA---RGS 145
ET ES+ T + KH K + G + + + AA +R + ++ S + R
Sbjct: 106 ETETESEATSTTWSLGKHTQIIGLSEKLINGNNLSAKIEAAREIRKMVRKSSSSSKTRAK 285
Query: 146 LAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPC 205
LA G I PLV ML S +L ++ SL ALLNL + N+ NK IV GA+ +++L++
Sbjct: 286 LAAAGVIEPLVLMLSSSNLDARQSSLLALLNLAVRNERNKVKIVTDGAMPPLVELLK--- 456
Query: 206 VVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRA 265
+ +S + E A L LSA SNKPII +SGA P LV+ILK+ S Q K DA+ A
Sbjct: 457 MQNSGIRELATAAILTLSAAASNKPIIAASGAAPLLVQILKS------GSVQGKVDAVTA 618
Query: 266 LYNLSINQTNISFVLETDLVVFLINSIED----MEVSERVLSILSNLVSSPEGRKAISAV 321
L+NLS + N +L+ V L+N +++ + +E+ ++L L +S EGR AIS
Sbjct: 619 LHNLSTSIANSIELLDASAVFPLLNLLKECKKYSKFAEKATALLEILSNSEEGRTAISIA 798
Query: 322 KDAITVLVDVL 332
I LV+ +
Sbjct: 799 DGGILTLVETV 831
>CO982694
Length = 840
Score = 120 bits (300), Expect = 2e-27
Identities = 78/231 (33%), Positives = 123/231 (52%)
Frame = -3
Query: 265 ALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKDA 324
AL+NLS N ++ + +V L++ E+SE+ L+ L NL + G+KAI
Sbjct: 835 ALHNLSTVLENACPLVSSGVVPILLDVSSIKEISEKALATLGNLSVTLMGKKAIENNSMV 656
Query: 325 ITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQK 384
+++L+W D P+CQE + YILMI+AH++ R+ M +AGIV LLE+ L+G+ LAQK
Sbjct: 655 PETFIEILSWEDKPKCQELSVYILMILAHQSSLQRKKMAQAGIVPVLLEVVLLGSPLAQK 476
Query: 385 RASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKTDGGGKECLMEEVNMMS 444
RA ++LQ F+ ++ +V + +P+ +
Sbjct: 475 RAMKLLQWFKDERQTKVGPHSGPQTPRFAMGSPVNQREA--------------------K 356
Query: 445 DEKKAVKQLVQQSLQNNMMKIVKRANLRQDFVPSERFASLTSSSTSKSLPF 495
+ K+ +K LV+QSL NM I +RAN + S R SL S++SKSLP+
Sbjct: 355 EGKRLMKSLVKQSLHRNMEIIAQRANAAGE---SSRLKSLIISTSSKSLPY 212
>TC232253
Length = 552
Score = 115 bits (287), Expect = 6e-26
Identities = 61/86 (70%), Positives = 66/86 (75%)
Frame = +1
Query: 410 LGLTVSAPICASSSSLVKTDGGGKECLMEEVNMMSDEKKAVKQLVQQSLQNNMMKIVKRA 469
LG VSAPIC SSS+ GGG+E E+ MMS+EKKAVKQLVQQSLQNNM KIVKRA
Sbjct: 7 LGAAVSAPICGSSSASPDGGGGGREFFEEDEEMMSEEKKAVKQLVQQSLQNNMRKIVKRA 186
Query: 470 NLRQDFVPSERFASLTSSSTSKSLPF 495
NL D PS+ F SLTSSSTSKSLPF
Sbjct: 187 NLPHDIAPSDHFMSLTSSSTSKSLPF 264
>TC228634 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g67530
{Arabidopsis thaliana;} , partial (35%)
Length = 1251
Score = 109 bits (272), Expect = 3e-24
Identities = 86/305 (28%), Positives = 159/305 (51%), Gaps = 7/305 (2%)
Frame = +2
Query: 97 NAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLV 156
+A+ + E+ + +K + + G + K+ ++R L ++D EAR + G + L+
Sbjct: 101 SAQEEDSEQYFSFLKVLTE---GNNWRKQCEVVEQLRLLLRDDEEARIFMGANGFVEALL 271
Query: 157 GMLDSEDLHSQIDSL----YALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVS 212
L S + +L AL NL + N+ NK ++ G +L L+E SS
Sbjct: 272 QFLQSAVREGSLMALESGAMALFNLAVNNNRNKEIMLSAG----VLSLLEEMISKTSSYG 439
Query: 213 EAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSIN 272
A +L LS L+ KP+IG + A+ FL+++L+ S S Q KQD+L ALYNLS
Sbjct: 440 -CTTALYLNLSCLEEAKPMIGVTQAVQFLIQLLQ-----SDSDVQCKQDSLHALYNLSTV 601
Query: 273 QTNISFVLETDLVVFLINSI---EDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLV 329
+NI +L ++ L + + D +E+ +++L NL +S GR+ I + I L
Sbjct: 602 PSNIPCLLSFGIISGLQSLLVGEGDSIWTEKCVAVLINLATSQVGREEIVSTPGLIGALA 781
Query: 330 DVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRI 389
+L+ + E QE+A L+I+ +++ + +++ G++ +L+ +++ GT Q++A ++
Sbjct: 782 SILDTGELIE-QEQAVSCLLILCNRSEECSEMVLQEGVIPALVSISVNGTPRGQEKAQKL 958
Query: 390 LQCFR 394
L FR
Sbjct: 959 LMLFR 973
>TC209517 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g67530
{Arabidopsis thaliana;} , partial (34%)
Length = 1350
Score = 106 bits (264), Expect = 3e-23
Identities = 83/271 (30%), Positives = 145/271 (52%), Gaps = 7/271 (2%)
Frame = +1
Query: 131 RVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSL----YALLNLGIANDANKA 186
++R L ++D EAR + G + L+ L S + +L AL NL + N+ NK
Sbjct: 85 QLRLLLRDDEEARIFMGTNGFVEALMQFLQSAVHEANAMALENGAMALFNLAVNNNRNKE 264
Query: 187 AIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILK 246
++ G +L L+E SS A VA +L LS LD K +IG+S A+ FL++IL+
Sbjct: 265 IMIATG----ILSLLEEMISKTSSYGCA-VALYLNLSCLDKAKHMIGTSQAVQFLIQILE 429
Query: 247 NLDNSSKSSSQVKQDALRALYNLSINQTNISFVLET---DLVVFLINSIEDMEVSERVLS 303
+K+ Q K D+L ALYNLS +NI +L + D + L+ D +E+ ++
Sbjct: 430 -----AKTEVQCKIDSLHALYNLSTVPSNIPNLLSSGIMDGLQSLLVDQGDCMWTEKCIA 594
Query: 304 ILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMI 363
+L NL GR+ + I+ L L+ T P QE+A+ L+I+ +++ Q ++
Sbjct: 595 VLINLAVYQAGREKMMLAPGLISALASTLD-TGEPIEQEQAASCLLILCNRSEECCQMVL 771
Query: 364 EAGIVSSLLELTLVGTALAQKRASRILQCFR 394
+ G++ +L+ +++ GT+ +++A ++L FR
Sbjct: 772 QEGVIPALVSISVNGTSRGREKAQKLLMVFR 864
>TC206834 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, partial (89%)
Length = 1740
Score = 102 bits (255), Expect = 3e-22
Identities = 79/278 (28%), Positives = 138/278 (49%), Gaps = 2/278 (0%)
Frame = +3
Query: 110 MKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQID 169
++ +V DL ++ AA +R L K E R +A GAI PL+ ++ S DL Q
Sbjct: 231 IRQLVADLHSSSIDDQKQAAMEIRLLAKNKPENRIKIAKAGAIKPLISLISSPDLQLQEY 410
Query: 170 SLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNK 229
+ A+LNL + D NK I GA+ +++ + S ++ E L LS ++ NK
Sbjct: 411 GVTAILNLSLC-DENKEVIASSGAIKPLVRALNSG---TATAKENAACALLRLSQVEENK 578
Query: 230 PIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLI 289
IG SGAIP LV +L++ + K+DA ALY+L + N ++ ++ L+
Sbjct: 579 AAIGRSGAIPLLVSLLES------GGFRAKKDASTALYSLCTVKENKIRAVKAGIMKVLV 740
Query: 290 NSIEDME--VSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYI 347
+ D E + ++ ++S LV+ PE R A+ + + VLV+++ + +E I
Sbjct: 741 ELMADFESNMVDKSAYVVSVLVAVPEARVAL-VEEGGVPVLVEIVE-VGTQRQKEIVVVI 914
Query: 348 LMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKR 385
L+ + + A R + G + L+ L+ GT A+++
Sbjct: 915 LLQVCEDSVAYRTMVAREGAIPPLVSLSQSGTNRAKQK 1028
>TC218468
Length = 992
Score = 83.2 bits (204), Expect = 2e-16
Identities = 59/189 (31%), Positives = 101/189 (53%)
Frame = +2
Query: 79 EEKTEKLLDLLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKE 138
E E+ L N ++ +++ + + ++ +V+ L +RR A +RSL+K
Sbjct: 455 EHNIEQPTGLTNGKLKKSDGSFRDVTGDIAAIEALVRKLSCRSVEERRAAVTELRSLSKR 634
Query: 139 DSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKML 198
++ R +A GAI LV +L SED+ +Q +++ ++LNL I + NK I+ GA+ ++
Sbjct: 635 STDNRILIAEAGAIPVLVNLLTSEDVLTQDNAVTSILNLSIYEN-NKGLIMLAGAIPSIV 811
Query: 199 KLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQV 258
+++ + + E A LS D NK IIG+SGAIP LV +L+N S +
Sbjct: 812 QVLRAGTM---EARENAAATLFSLSLADENKIIIGASGAIPALVELLQN------GSPRG 964
Query: 259 KQDALRALY 267
K+DA AL+
Sbjct: 965 KKDAATALF 991
>TC210964 similar to UP|Q84TU4 (Q84TU4) Arm repeat-containing protein,
partial (22%)
Length = 608
Score = 78.6 bits (192), Expect = 6e-15
Identities = 54/151 (35%), Positives = 83/151 (54%)
Frame = +2
Query: 88 LLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLA 147
+++ V ET A+ E T+++++V+ LR D +R A A +R L K + + R ++A
Sbjct: 176 IVSSPVVETRADLSAIE---TQVRNLVEGLRSSDVDTQREATAELRLLAKHNMDNRIAIA 346
Query: 148 MLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVV 207
GAI+ LV +L S D Q +++ ALLNL I ND NK AI GA+ ++ ++E+
Sbjct: 347 NCGAINLLVDLLQSTDTTIQENAVTALLNLSI-NDNNKTAIANAGAIEPLIHVLETG--- 514
Query: 208 DSSVSEAIVANFLGLSALDSNKPIIGSSGAI 238
E A LS ++ NK IG SGAI
Sbjct: 515 SPEAKENSAATLFSLSVIEENKIFIGRSGAI 607
Score = 56.6 bits (135), Expect = 2e-08
Identities = 41/140 (29%), Positives = 73/140 (51%), Gaps = 1/140 (0%)
Frame = +2
Query: 141 EARGSLAMLGA-ISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLK 199
E R L+ + + LV L S D+ +Q ++ L L N N+ AI GA++ ++
Sbjct: 197 ETRADLSAIETQVRNLVEGLRSSDVDTQREATAELRLLAKHNMDNRIAIANCGAINLLVD 376
Query: 200 LIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVK 259
L++S D+++ E V L LS D+NK I ++GAI L+ +L+ S + K
Sbjct: 377 LLQS---TDTTIQENAVTALLNLSINDNNKTAIANAGAIEPLIHVLET------GSPEAK 529
Query: 260 QDALRALYNLSINQTNISFV 279
+++ L++LS+ + N F+
Sbjct: 530 ENSAATLFSLSVIEENKIFI 589
>TC210094 weakly similar to UP|Q6K5K9 (Q6K5K9) Avr9/Cf-9 rapidly elicited
protein-like, partial (16%)
Length = 1020
Score = 72.4 bits (176), Expect = 4e-13
Identities = 71/279 (25%), Positives = 131/279 (46%), Gaps = 4/279 (1%)
Frame = +2
Query: 152 ISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSV 211
+SPL DLH D + +LNL I +D K+ + ++ ++ + S
Sbjct: 50 LSPLAAASTDPDLHE--DLITTVLNLSIHDDNKKSFAEDPALISLLIDALKCGTIQTRSN 223
Query: 212 SEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSI 271
+ A + LSA+DSNK IIG SGAI L+ +L + +DA A++NL +
Sbjct: 224 AAATIFT---LSAIDSNKHIIGESGAIKHLLELL------DEGQPFAMKDAASAIFNLCL 376
Query: 272 NQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDV 331
N + V ++N + D + + +L+IL+ L S P+ + + DA+ +L+ +
Sbjct: 377 VHENKGRTVRDGAVRVILNKMMDHILVDELLAILALLSSHPKAVEEMGDF-DAVPLLLGI 553
Query: 332 LNWTDSPECQEKASYILMIMAHKAYADR----QAMIEAGIVSSLLELTLVGTALAQKRAS 387
+ + S +E IL + ++DR + E +L +L GT+ A+++A+
Sbjct: 554 IRESTSERSKENCVAILYTI---CFSDRTKLKEIREEEKANGTLTKLGKCGTSRAKRKAN 724
Query: 388 RILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLV 426
IL+ RL++ ++ + +P+ +SS LV
Sbjct: 725 GILE--RLNRSPSLTHTA*RN*GSRIRKSPLFVNSSFLV 835
>TC216726 similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing protein
homolog, partial (35%)
Length = 1115
Score = 70.1 bits (170), Expect = 2e-12
Identities = 63/224 (28%), Positives = 105/224 (46%), Gaps = 40/224 (17%)
Frame = +3
Query: 208 DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKN------------------LD 249
DS E V L LS ++NK I SSGA+P +V +LK +D
Sbjct: 6 DSRTQEHAVTALLNLSIYENNKGSIVSSGAVPGIVHVLKKGSMEARENAAATLFSLSVID 185
Query: 250 NS-----------------SKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSI 292
+ S+ + + K+DA AL+NL I Q N + ++ L+ +
Sbjct: 186 ENKVTIGSLGAIPPLVTLLSEGNQRGKKDAATALFNLCIYQGNKGKAVRAGVIPTLMRLL 365
Query: 293 EDME--VSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMI 350
+ + + L+IL+ L S PEG+ I A +A+ VLV+ + SP +E A+ +L+
Sbjct: 366 TEPSGGMVDEALAILAILASHPEGKATIRA-SEAVPVLVEFIG-NGSPRNKENAAAVLV- 536
Query: 351 MAHKAYADRQAMIEA---GIVSSLLELTLVGTALAQKRASRILQ 391
H D+Q + +A G++ LLEL GT +++A ++L+
Sbjct: 537 --HLCSGDQQYLAQAQELGVMGPLLELAQNGTDRGKRKAGQLLE 662
Score = 56.6 bits (135), Expect = 2e-08
Identities = 56/182 (30%), Positives = 87/182 (47%)
Frame = +3
Query: 88 LLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLA 147
LLN+ ++E N S + + HV+K E R AAA + SL+ D E + ++
Sbjct: 39 LLNLSIYENNKGSIVSSGAVPGIVHVLKKGSME---ARENAAATLFSLSVID-ENKVTIG 206
Query: 148 MLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVV 207
LGAI PLV +L + + D+ AL NL I NK V+ G + +++L+ P
Sbjct: 207 SLGAIPPLVTLLSEGNQRGKKDAATALFNLCI-YQGNKGKAVRAGVIPTLMRLLTEPS-- 377
Query: 208 DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALY 267
V EA+ A L++ K I +S A+P LV + N S + K++A L
Sbjct: 378 GGMVDEAL-AILAILASHPEGKATIRASEAVPVLVEFIGN------GSPRNKENAAAVLV 536
Query: 268 NL 269
+L
Sbjct: 537 HL 542
>TC222105
Length = 904
Score = 68.2 bits (165), Expect = 8e-12
Identities = 61/217 (28%), Positives = 107/217 (49%), Gaps = 2/217 (0%)
Frame = +1
Query: 180 ANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIP 239
A +A + V+ G V ++ +++ + S A A F L+ D NK IG GA+
Sbjct: 1 AREAEQGEDVRSGFVPFLIDVLKGG--LGESQEHAAGALF-SLALDDDNKMAIGVLGALH 171
Query: 240 FLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSE 299
L+ L+ S + + D+ ALY+LS+ Q+N +++ +V L++ + ++
Sbjct: 172 PLMHALR------AESERTRHDSALALYHLSLVQSNRMKLVKLGVVPTLLSMVVAGNLAS 333
Query: 300 RVLSILSNLVSSPEGRKAISAVKDAITVLVDVL--NWTDSPECQEKASYILMIMAHKAYA 357
RVL IL NL EGR A+ +A+ +LV +L N DS +E L ++H++
Sbjct: 334 RVLLILCNLAVCTEGRTAMLDA-NAVEILVSLLRGNELDSEATRENCVAALYALSHRSLR 510
Query: 358 DRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFR 394
+ EA + L E+ GT A+++A ++L R
Sbjct: 511 FKGLAKEARVAEVLKEIEETGTERAREKARKVLHMLR 621
>BM309421 similar to GP|28974687|gb arm repeat-containing protein {Nicotiana
tabacum}, partial (17%)
Length = 433
Score = 60.8 bits (146), Expect = 1e-09
Identities = 45/148 (30%), Positives = 81/148 (54%), Gaps = 1/148 (0%)
Frame = +3
Query: 186 AAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRIL 245
A I++ GA+ ++ L+E + E A LS +D+NK IG SGA+ LV +L
Sbjct: 3 ALIMEAGAIEPLIHLLEKG---NDGAKENSAAALFSLSVIDNNKAKIGRSGAVKALVGLL 173
Query: 246 KNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDME-VSERVLSI 304
+ + + K+DA AL+NLSI N + +++ V FL+ ++ + + ++ +++
Sbjct: 174 ------ASGTLRGKKDAATALFNLSIFHENKARIVQAGAVKFLVLLLDPTDKMVDKAVAL 335
Query: 305 LSNLVSSPEGRKAISAVKDAITVLVDVL 332
L+NL + EGR I A + I LV+++
Sbjct: 336 LANLSTIAEGRIEI-AREGGIPSLVEIV 416
Score = 56.2 bits (134), Expect = 3e-08
Identities = 47/130 (36%), Positives = 68/130 (52%)
Frame = +3
Query: 118 RGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNL 177
+G D K AAA ++++A+ + GA+ LVG+L S L + D+ AL NL
Sbjct: 54 KGNDGAKENSAAALFSLSVIDNNKAK--IGRSGAVKALVGLLASGTLRGKKDAATALFNL 227
Query: 178 GIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGA 237
I ++ NKA IV+ GAV K L L+ P D V +A VA LS + + I G
Sbjct: 228 SIFHE-NKARIVQAGAV-KFLVLLLDP--TDKMVDKA-VALLANLSTIAEGRIEIAREGG 392
Query: 238 IPFLVRILKN 247
IP LV I+++
Sbjct: 393 IPSLVEIVES 422
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/100 (35%), Positives = 53/100 (53%)
Frame = +3
Query: 150 GAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDS 209
GAI PL+ +L+ + ++ +S AL +L + D NKA I + GAV ++ L+ S +
Sbjct: 21 GAIEPLIHLLEKGNDGAKENSAAALFSLSVI-DNNKAKIGRSGAVKALVGLLASGTL--R 191
Query: 210 SVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLD 249
+A A F LS NK I +GA+ FLV +L D
Sbjct: 192 GKKDAATALF-NLSIFHENKARIVQAGAVKFLVLLLDPTD 308
>BU765405
Length = 421
Score = 60.8 bits (146), Expect = 1e-09
Identities = 44/131 (33%), Positives = 70/131 (52%), Gaps = 2/131 (1%)
Frame = +1
Query: 100 SKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGML 159
S + E T ++ ++ L+ D R AA +R LTK R L A++PLV ML
Sbjct: 49 SPEAEATAVAVRRALELLQLNDPVLRVQAARDIRRLTKTSQRCRRQLRF--AVAPLVSML 222
Query: 160 --DSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVA 217
DS + H +L ALLNL + ++ NK +IV+ GA+ ++ ++SP + ++ E A
Sbjct: 223 RVDSSEFHEP--ALLALLNLAVQDEKNKISIVEAGALEPIISFLKSP---NPNLQEYATA 387
Query: 218 NFLGLSALDSN 228
+ L LSA +N
Sbjct: 388 SLLTLSASPTN 420
>BG156589 similar to GP|12483839|gb| p6.9 {Heliocoverpa armigera
nucleopolyhedrovirus G4}, partial (16%)
Length = 236
Score = 57.4 bits (137), Expect = 1e-08
Identities = 40/83 (48%), Positives = 51/83 (61%), Gaps = 4/83 (4%)
Frame = +1
Query: 12 IVLHHRHHAKNTTA-GNFRFSASSFRRIIFDALSCGGASRHHRHYHREEEIS---SVAST 67
I L HRH AKN TA G+FR RR+IFDA+SCGG SR +R+ R ++S +S
Sbjct: 1 IALDHRHSAKNATAAGHFRV----LRRLIFDAVSCGGTSR-YRYRQRGGDVSDGGDFSSA 165
Query: 68 ATVKELRGEVQEEKTEKLLDLLN 90
A E +EE++EKL DLLN
Sbjct: 166ACHTE-----KEERSEKLSDLLN 219
>BQ253474
Length = 276
Score = 55.8 bits (133), Expect = 4e-08
Identities = 28/58 (48%), Positives = 40/58 (68%), Gaps = 7/58 (12%)
Frame = +2
Query: 1 MAKCHRSNVDSIVL----HHRHHAKNTTAGNFRF---SASSFRRIIFDALSCGGASRH 51
MA HR+NV+++VL HH H +K+ + F +A+SFRRIIFDA+SCG +SR+
Sbjct: 50 MANYHRNNVENVVLGRHHHHSHRSKSASVAGHTFRLWNAASFRRIIFDAVSCGASSRY 223
>TC216817 weakly similar to UP|KHLX_HUMAN (Q9Y2M5) Kelch-like protein X,
partial (5%)
Length = 1071
Score = 54.7 bits (130), Expect = 9e-08
Identities = 70/291 (24%), Positives = 128/291 (43%), Gaps = 5/291 (1%)
Frame = +1
Query: 117 LRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLN 176
LR ED+ A + +L + + + GA+ P++G+L S SQ ++ L
Sbjct: 25 LRSEDAGVHYEAVGVIGNLVHSSPNIKKEVLLAGALQPVIGLLSSCCSESQREAALLLGQ 204
Query: 177 LGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALD-SNKPIIGSS 235
+ K IV+ GAV ++++++SP V +S A LG A D N+ I +
Sbjct: 205 FAATDSDCKVHIVQRGAVQPLIEMLQSPDVQLREMS----AFALGRLAQDPHNQAGIAHN 372
Query: 236 GAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSIEDM 295
G LV +LK LD+ + S ++ +A ALY L+ N+ N S + + ++ +
Sbjct: 373 GG---LVPLLKLLDSKNGS---LQHNAAFALYGLADNEDNASDFIR-------VGGVQRL 513
Query: 296 EVSERVLSILSNLVSSPEGR---KAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMA 352
+ E ++ + V+ R K V + + L+ V CQ + + + +A
Sbjct: 514 QDGEFIVQATKDCVAKTLKRLEEKIHGRVLNHLLYLMRV----SEKGCQRR---VALALA 672
Query: 353 HKAYADRQAMIEAGIVSSLLELTLVGTALA-QKRASRILQCFRLDKGKQVS 402
H +D Q +I L + L+G++ + Q+ + C DK +S
Sbjct: 673 HLCSSDDQRIIFIDHYGLELLIGLLGSSSSKQQLDGAVALCKLADKASTLS 825
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.129 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,418,232
Number of Sequences: 63676
Number of extensions: 208642
Number of successful extensions: 1539
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1441
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1490
length of query: 495
length of database: 12,639,632
effective HSP length: 101
effective length of query: 394
effective length of database: 6,208,356
effective search space: 2446092264
effective search space used: 2446092264
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC133571.8


