

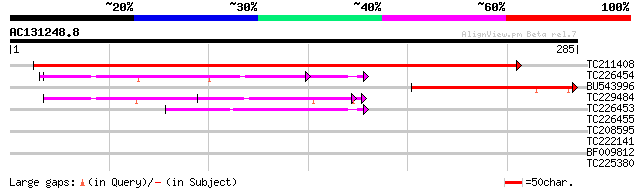
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131248.8 + phase: 0
(285 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211408 similar to UP|Q8L4J8 (Q8L4J8) CTP:phosphorylcholine cyt... 437 e-123
TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 108 2e-24
BU543996 similar to GP|21668498|dbj CTP:phosphorylcholine cytidy... 96 2e-20
TC229484 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytid... 80 1e-15
TC226453 weakly similar to UP|Q84JV7 (Q84JV7) CTP:ethanolamine c... 49 3e-06
TC226455 homologue to UP|Q84JV7 (Q84JV7) CTP:ethanolamine cytidy... 39 0.002
TC208595 weakly similar to UP|Q9LIQ0 (Q9LIQ0) Arabidopsis thalia... 38 0.006
TC222141 homologue to UP|HMGI_HUMAN (P17096) High mobility group... 28 3.6
BF009812 28 4.7
TC225380 27 8.1
>TC211408 similar to UP|Q8L4J8 (Q8L4J8) CTP:phosphorylcholine
cytidylyltransferase , partial (83%)
Length = 870
Score = 437 bits (1125), Expect = e-123
Identities = 205/245 (83%), Positives = 226/245 (91%)
Frame = +2
Query: 13 DKPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQERY 72
D+PVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCC+D++THKYKGKTVM E ERY
Sbjct: 89 DRPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCNDQVTHKYKGKTVMTEAERY 268
Query: 73 ESLRHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKF 132
ESLRHCKWVDEVIPD PWVINQEF+DKH IDYVAHD+LPYAD +GA NDVYEFVK++G+F
Sbjct: 269 ESLRHCKWVDEVIPDAPWVINQEFLDKHNIDYVAHDSLPYADANGAANDVYEFVKSVGRF 448
Query: 133 KETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQE 192
KETKRTEGISTSD+IMRI+KDYNQYV+RNLDRGYSR ELGVSYVKEKRLR+N LK LQE
Sbjct: 449 KETKRTEGISTSDVIMRIVKDYNQYVLRNLDRGYSRNELGVSYVKEKRLRVNRRLKTLQE 628
Query: 193 RVKKQQETVGKKIGTVRRIAGMNRTEWVENADRLVAGFLEMFEEGCHKMGTAIRDRIQEQ 252
+VK+ QE VG+KI V + AGM+R EWVENADR VAGFLEMFEEGCHKMGTAIRDRIQE+
Sbjct: 629 KVKEHQEKVGEKIQIVAKTAGMHRNEWVENADRWVAGFLEMFEEGCHKMGTAIRDRIQER 808
Query: 253 LKAQQ 257
L+ QQ
Sbjct: 809 LRGQQ 823
>TC226454 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (71%)
Length = 1889
Score = 108 bits (271), Expect = 2e-24
Identities = 60/140 (42%), Positives = 84/140 (59%), Gaps = 4/140 (2%)
Frame = +1
Query: 16 VRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQERYESL 75
+RVY DG +DL H+GHA +L QAK L+VG SDE KG V++ +ER +
Sbjct: 421 IRVYMDGCFDLMHYGHANALRQAKAL--GDELVVGLVSDEEIVANKGPPVLSMEERLALV 594
Query: 76 RHCKWVDEVIPDVPWVINQEFIDK----HKIDYVAHDALPYADTSGAGNDVYEFVKAIGK 131
KWVDEVI D P+ I ++F+++ +KIDYV H P G D Y K G+
Sbjct: 595 SGLKWVDEVITDAPYAITEQFLNRLFHEYKIDYVIHGDDPCLLPD--GTDAYAAAKKAGR 768
Query: 132 FKETKRTEGISTSDIIMRII 151
+K+ KRTEG+S++DI+ RI+
Sbjct: 769 YKQIKRTEGVSSTDIVGRIL 828
Score = 88.2 bits (217), Expect = 4e-18
Identities = 56/165 (33%), Positives = 86/165 (51%), Gaps = 2/165 (1%)
Frame = +1
Query: 18 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGK--TVMNEQERYESL 75
VY DG +DLFH GH L++A++ +LLVG SDE +++G +M+ ER S+
Sbjct: 994 VYIDGAFDLFHAGHVEILKRAREL--GDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSV 1167
Query: 76 RHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKFKET 135
C++VDEVI PW I + I I V H + T D YE K++G F
Sbjct: 1168 LACRYVDEVIIGSPWEITNDMITTFNISVVVHGTVA-EKTLNCELDPYEVPKSMGIFHLL 1344
Query: 136 KRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKR 180
+ + I+T+ + RI+ ++ YV RN + S + Y +EK+
Sbjct: 1345 ESPKDITTATVAQRIMANHEAYVKRNAKKTRSEQR----YYEEKK 1467
>BU543996 similar to GP|21668498|dbj CTP:phosphorylcholine
cytidylyltransferase {Arabidopsis thaliana}, partial
(21%)
Length = 613
Score = 95.9 bits (237), Expect = 2e-20
Identities = 51/96 (53%), Positives = 61/96 (63%), Gaps = 13/96 (13%)
Frame = -1
Query: 203 KKIGTVRRIAGMNRTEWVENADRLVAGFLEMFEEGCHKMGTAIRDRIQEQLKAQQLKSLL 262
+KI V + GM R EWVENADR VAGFLEMFEEGCHKMG IRDRIQE+L+ QQ +
Sbjct: 613 EKIQIVAKTXGMXRNEWVENADRWVAGFLEMFEEGCHKMGXXIRDRIQERLRGQQSRDGT 434
Query: 263 Y------DEWDDDVDDEFYEDES-------VEYYSD 285
D+ DDD ++ +Y+DE EYY D
Sbjct: 433 LRLQNGEDDKDDDDEEYYYDDEEDSDEEYFEEYYDD 326
>TC229484 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (61%)
Length = 947
Score = 80.1 bits (196), Expect = 1e-15
Identities = 55/167 (32%), Positives = 84/167 (49%), Gaps = 5/167 (2%)
Frame = +2
Query: 18 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKG--KTVMNEQERYESL 75
VY DG +DLFH GH L A+ +LLVG +D+ +G + +MN ER S+
Sbjct: 374 VYIDGAFDLFHAGHVEILRLARDL--GDFLLVGIHTDQTVSATRGSHRPIMNLHERSLSV 547
Query: 76 RHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYA-DTSGAGNDVYEFVKAIGKFKE 134
C++VDEVI PW I+++ + I V H + + D + Y ++G FK
Sbjct: 548 LACRYVDEVIIGAPWEISKDMLTTFNISLVVHGTIAESNDFKKEECNPYAVPISMGIFKV 727
Query: 135 TKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKEL--GVSYVKEK 179
+ I+T+ II RI+ ++ Y RN + S K G S+V E+
Sbjct: 728 LESPLDITTTTIIRRIVCNHEAYQNRNKKKDESEKRYYEGKSHVSEE 868
Score = 48.5 bits (114), Expect = 3e-06
Identities = 31/92 (33%), Positives = 49/92 (52%), Gaps = 12/92 (13%)
Frame = +2
Query: 95 EFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKFKETKRTEGISTSDIIMRII--- 151
+ D++KID++ H P G D Y K G++K+ KRTEG+S++DI+ R++
Sbjct: 2 KLFDEYKIDFIIHGDDPCVLPDGT--DAYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCV 175
Query: 152 -------KDYN-QYVMRNLDRGYSRK-ELGVS 174
K++N + R G+S K E GVS
Sbjct: 176 RERSIAEKNHNHSSLQRQFSNGHSPKFEAGVS 271
>TC226453 weakly similar to UP|Q84JV7 (Q84JV7) CTP:ethanolamine
cytidylyltransferase (CTP-phosphoethanolamine
cytidylyltransferase), partial (11%)
Length = 620
Score = 48.5 bits (114), Expect = 3e-06
Identities = 30/102 (29%), Positives = 50/102 (48%)
Frame = +2
Query: 79 KWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKFKETKRT 138
++VDEVI PW I + I I V H + + D YE K++G F+ +
Sbjct: 2 RYVDEVIIGSPWEITNDMITTFNISVVVHGTVS-EKSLNCELDPYEIPKSMGIFRLLESP 178
Query: 139 EGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKR 180
+ I+T+ + RI+ ++ YV RN + S + Y +EK+
Sbjct: 179 KDITTATVAQRIMANHEAYVKRNAKKTRSEQR----YYEEKK 292
>TC226455 homologue to UP|Q84JV7 (Q84JV7) CTP:ethanolamine
cytidylyltransferase (CTP-phosphoethanolamine
cytidylyltransferase), partial (7%)
Length = 593
Score = 39.3 bits (90), Expect = 0.002
Identities = 15/24 (62%), Positives = 19/24 (78%)
Frame = +1
Query: 16 VRVYADGIYDLFHFGHARSLEQAK 39
+RVY DG +DL H+GHA +L QAK
Sbjct: 496 IRVYMDGCFDLMHYGHANALRQAK 567
>TC208595 weakly similar to UP|Q9LIQ0 (Q9LIQ0) Arabidopsis thaliana genomic
DNA, chromosome 3, BAC clone:F14O13, partial (20%)
Length = 979
Score = 37.7 bits (86), Expect = 0.006
Identities = 34/116 (29%), Positives = 55/116 (47%), Gaps = 7/116 (6%)
Frame = +1
Query: 110 LPYADTSGAGNDVYEFVKAIGKFKETKRTE-GISTSDIIMRIIKDYNQYVMRNLDRGYSR 168
L Y DT G +K K+ +TK + G+S S+I++ KD+NQYV Y
Sbjct: 289 LDYEDTIGD-------LKTRFKYAKTKPSRFGLSASEILLMDDKDFNQYVSLKKLAPYWE 447
Query: 169 KELGVSYVKEKRLRMNMGLKK------LQERVKKQQETVGKKIGTVRRIAGMNRTE 218
+E +S K+KR + M K+ L ++ KK + GK + + N+T+
Sbjct: 448 EEWKLS--KQKRYMLKMRAKELLCASSLDKKKKKSKVDSGKITSSKSVVENKNQTQ 609
>TC222141 homologue to UP|HMGI_HUMAN (P17096) High mobility group protein
HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook 1),
partial (11%)
Length = 564
Score = 28.5 bits (62), Expect = 3.6
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = -1
Query: 106 AHDALPYADTSGAGNDVYEFVKAIGKFKE 134
AHDA P ++G G D + V A+G F +
Sbjct: 459 AHDANPSLSSAGEGGDAIDDVAAVGDFHD 373
>BF009812
Length = 247
Score = 28.1 bits (61), Expect = 4.7
Identities = 11/21 (52%), Positives = 18/21 (85%)
Frame = +2
Query: 131 KFKETKRTEGISTSDIIMRII 151
++K+ KRTEG+S++DI RI+
Sbjct: 2 RYKQIKRTEGVSSTDIEGRIL 64
>TC225380
Length = 768
Score = 27.3 bits (59), Expect = 8.1
Identities = 15/36 (41%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Frame = +1
Query: 247 DRIQEQLKAQQLKSLL-YDEWDDDVDDEFYEDESVE 281
D I + A+Q KS D+ DDD DD+F +DE ++
Sbjct: 52 DTITLRKLAEQAKSFRPNDDDDDDSDDDFSDDEELQ 159
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,374,624
Number of Sequences: 63676
Number of extensions: 120792
Number of successful extensions: 693
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 662
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 685
length of query: 285
length of database: 12,639,632
effective HSP length: 96
effective length of query: 189
effective length of database: 6,526,736
effective search space: 1233553104
effective search space used: 1233553104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC131248.8


