

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.9 + phase: 0 /pseudo
(311 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
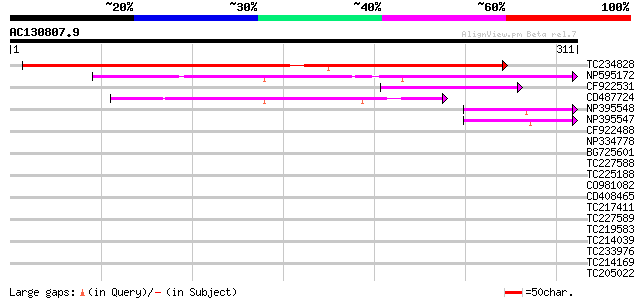
Score E
Sequences producing significant alignments: (bits) Value
TC234828 228 3e-60
NP595172 polyprotein [Glycine max] 98 4e-21
CF922531 56 2e-08
CD487724 54 1e-07
NP395548 reverse transcriptase [Glycine max] 49 3e-06
NP395547 reverse transcriptase [Glycine max] 47 1e-05
CF922488 40 0.001
NP334778 reverse transcriptase [Glycine max] 39 0.002
BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] -... 38 0.007
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 32 0.48
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 30 1.4
CO981082 30 1.4
CD408465 30 1.8
TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%) 30 1.8
TC227589 30 1.8
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 29 2.4
TC214039 homologue to UP|Q9M4R0 (Q9M4R0) Ubiquitin conjugating p... 29 2.4
TC233976 29 3.1
TC214169 similar to UP|Q8LF25 (Q8LF25) DNA-damage inducible prot... 29 3.1
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 29 3.1
>TC234828
Length = 857
Score = 228 bits (581), Expect = 3e-60
Identities = 118/270 (43%), Positives = 169/270 (61%), Gaps = 4/270 (1%)
Frame = +3
Query: 8 NCNEEGHIGSQCKQPKRAPTTGRVFAMAGTQTENEDRLIRGTCYINNTPLVAIIDTGATH 67
N G++ + R RVFAM+G++ D LIRG C I + L + D+GATH
Sbjct: 66 NSTRSGNVSNNNISGGRPKVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATH 245
Query: 68 CFIAFDCVSALGLDLSDMNGEMVVETPTKGSVTTSLVCLRCPLSMFGHDFEMDLVCLHLS 127
FI+ CV LGL +++ +MVV TPT VTTS VCL+CP+ + G F DL+CL L+
Sbjct: 246 SFISHACVERLGLCATELPYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLA 425
Query: 128 GMDVILGMNWLEYNHVHINCFSKSVCFSSAEEESGA*FLSTKQLKQ----MERDGILMFS 183
+DVILGM+WL NH+ ++C K + F G + ++ LK+ E + + +
Sbjct: 426 HLDVILGMDWLSTNHIFLDCKEKMLVF-------GGDVVPSEPLKEDAANEETEDVRTYM 584
Query: 184 LMASLSIENQAMIDKL*VVCDFPEVLPDEIPDMPPKREVEFSIDLVPRTKPVSMAPYRMS 243
++ S+ +E A + + VV +FPEV PD++ ++PP+REVEF ID+VP PVS+APYRMS
Sbjct: 585 VLFSMYVEEDAEVSCIPVVSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMS 764
Query: 244 ASELAELKKRLEDLLENKFVRPSVSPWGAP 273
ELAE+K +++DLL +FVRPS SPWGAP
Sbjct: 765 PVELAEVKAQVQDLLSKQFVRPSASPWGAP 854
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 98.2 bits (243), Expect = 4e-21
Identities = 74/290 (25%), Positives = 133/290 (45%), Gaps = 24/290 (8%)
Frame = +1
Query: 46 IRGTCYINNTPLVAIIDTGATHCFIAFDCVSALGLDLSDMNGEMVVETPTKGSVTTSL-V 104
IR T + + ++D G++ FI L L + V+ G + ++ +
Sbjct: 1171 IRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAPNLRVLVG--NGQILSAEGI 1344
Query: 105 CLRCPLSMFGHDFEMDLVCLHLSGMDVILGMNWL--------EYNHVHINCFSKSVCFS- 155
+ PL + G + ++ + L +SG DVILG WL +Y + + F +
Sbjct: 1345 VQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADYAALTLKFFQNDKFITL 1524
Query: 156 SAEEESGA*FLSTKQLKQMERDGILMFSLMASLSIENQAMIDKL*VVCDFPEVLPDEIP- 214
E S A ++++ + L I+ + D L D P + E+
Sbjct: 1525 QGEGNSEATQAQLHHFRRLQNTKSIEECFAIQL-IQKEVPEDTL---KDLPTNIDPELAI 1692
Query: 215 -------------DMPPKREVEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENK 261
+PP+RE + +I L + PV + PYR ++ +++K ++++L
Sbjct: 1693 LLHTYAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQG 1872
Query: 262 FVRPSVSPWGAPVFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
++PS SP+ P+ LV KKDGS R C DYR LN +T+K+ +P+P +D+L+
Sbjct: 1873 IIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDELL 2022
>CF922531
Length = 602
Score = 56.2 bits (134), Expect = 2e-08
Identities = 31/79 (39%), Positives = 47/79 (59%), Gaps = 1/79 (1%)
Frame = -2
Query: 204 DFPEVLPDEIP-DMPPKREVEFSIDLVPRTKPVSMAPYRMSASELAELKKRLEDLLENKF 262
+F ++ P EIP +PP R +E IDLVPR + YR + E E++ ++++LLE +
Sbjct: 238 EFGDIFPKEIPLGLPPLRGIEHQIDLVPRASLPNRPTYRTNPQETKEIESQVKELLEKGW 59
Query: 263 VRPSVSPWGAPVFLVMKKD 281
V+ S+S V LV KKD
Sbjct: 58 VQESLSLCVVLVLLVPKKD 2
>CD487724
Length = 676
Score = 53.5 bits (127), Expect = 1e-07
Identities = 49/207 (23%), Positives = 90/207 (42%), Gaps = 22/207 (10%)
Frame = +3
Query: 56 PLVAIIDTGATHCFIAFDCVSALGLDLSDMNGEMVVETPTKGSVTTSLVCLRCPLSMFGH 115
PLV ++D G+TH F+ VS LGL + V + + +C P+S+
Sbjct: 21 PLVYLVDGGSTHNFVQQPLVSQLGLPCRS-TPPLRVMVGNGHHLKCTTICEAIPISIQNI 197
Query: 116 DFEMDLVCLHLSGMDVILGMNWL--------EYNHVHINCF-SKSVCFSSAEEESGA*FL 166
+F + L L + G +++LG+ WL +YN + + F + E E+ L
Sbjct: 198 EFLVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGLL 377
Query: 167 STKQLKQMERDGILMFSLMASLSIEN-------------QAMIDKL*VVCDFPEVLPDEI 213
+ QL+++ + + ++ EN Q ++D+ + P+
Sbjct: 378 NHHQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFSALFQ*PQ------ 539
Query: 214 PDMPPKREVEFSIDLVPRTKPVSMAPY 240
+PP RE + I L+P ++PV+M Y
Sbjct: 540 -GLPPARETDHHIHLLP*SEPVNMRLY 617
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 48.9 bits (115), Expect = 3e-06
Identities = 29/81 (35%), Positives = 44/81 (53%), Gaps = 19/81 (23%)
Frame = +1
Query: 250 LKKRLEDLLENKFVRP-SVSPWGAPVFLVMKKDG------------------SMRLCIDY 290
++K + LLE + P S S W +PV +V KK+G S +LCIDY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 291 RQLNKVTIKNRYPLPRIDDLM 311
R+LN+ T K+ +PLP +D ++
Sbjct: 181 RKLNEATRKDHFPLPFMDQML 243
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 46.6 bits (109), Expect = 1e-05
Identities = 29/81 (35%), Positives = 42/81 (51%), Gaps = 19/81 (23%)
Frame = +1
Query: 250 LKKRLEDLLENKFVRP-SVSPWGAPVFLVMKKDGSM------------------RLCIDY 290
++K + LLE + P S S W +PV +V KK G R+CIDY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 291 RQLNKVTIKNRYPLPRIDDLM 311
R+LN+ T K+ YPLP +D ++
Sbjct: 181 RKLNEATRKDHYPLPFMDQML 243
>CF922488
Length = 741
Score = 40.4 bits (93), Expect = 0.001
Identities = 16/35 (45%), Positives = 27/35 (76%)
Frame = +3
Query: 277 VMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLM 311
V+K+DG + +C+DYR LN + K+++PLP I+ L+
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLV 107
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 39.3 bits (90), Expect = 0.002
Identities = 15/27 (55%), Positives = 21/27 (77%)
Frame = +3
Query: 285 RLCIDYRQLNKVTIKNRYPLPRIDDLM 311
R+C+DYR LN+ + K+ +PLP ID LM
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILM 83
>BG725601 similar to PIR|H86337|H86 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (1%)
Length = 285
Score = 37.7 bits (86), Expect = 0.007
Identities = 17/37 (45%), Positives = 22/37 (58%)
Frame = -3
Query: 274 VFLVMKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDL 310
V +V K +G R+C DY LN K+ YPLP ID +
Sbjct: 124 VVMVKKPNGKWRICTDYIDLN*ACPKDAYPLPNIDHM 14
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 31.6 bits (70), Expect = 0.48
Identities = 28/113 (24%), Positives = 37/113 (31%), Gaps = 2/113 (1%)
Frame = +1
Query: 6 CFNCNEEGHIGSQCKQPKRAPTTGRVFAMAGTQTENEDRLIRGTCYINNTPLVAIIDTGA 65
CFNC EEGH C KR + CY+ + A
Sbjct: 61 CFNCGEEGHAAVNCSAVKR----------------------KKPCYV-----CGCLGHNA 159
Query: 66 THCFIAFDCVSALGLDLSDMNGEMVVETPTKGSVTTS--LVCLRCPLSMFGHD 116
C DC + G + P K + T+ +CL+C S GHD
Sbjct: 160 RQCSKVQDCF------ICKKGGHRAKDCPEKHTSTSKSIAICLKCGNS--GHD 294
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 30.0 bits (66), Expect = 1.4
Identities = 13/40 (32%), Positives = 20/40 (49%), Gaps = 1/40 (2%)
Frame = +1
Query: 6 CFNCNEEGHIGSQCK-QPKRAPTTGRVFAMAGTQTENEDR 44
C+ C E GHI CK PK+ GR + + ++ + R
Sbjct: 430 CYRCGERGHIEKNCKNSPKKLSQRGRRLSRSPVRSRSPRR 549
>CO981082
Length = 670
Score = 30.0 bits (66), Expect = 1.4
Identities = 17/58 (29%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Frame = +1
Query: 88 EMVVETPTKGSV-TTSLVCLRCPLSMFGHDFEMDLVCLHLSGMDVILGMNWLEYNHVH 144
E+ V +PTK T S++CL +SMF F ++C ++ +Y H H
Sbjct: 229 ELWVTSPTKAKAFTMSIICLCSCISMFPGHFPFSIICTVSEMQTNSETISTTKYKHTH 402
>CD408465
Length = 630
Score = 29.6 bits (65), Expect = 1.8
Identities = 20/58 (34%), Positives = 25/58 (42%), Gaps = 9/58 (15%)
Frame = -1
Query: 208 VLPDEIPD-MPPKREVEFSIDLVPRTKPVSMAPY--------RMSASELAELKKRLED 256
+ P EIP +PP R +E +DL+P S Y R A E KRL D
Sbjct: 630 IFPKEIPHGLPPSRGIEHQVDLLPGASLPSRPTYKSYPQETQRKDAHAKVEYVKRLYD 457
>TC217411 similar to UP|Q8L7S8 (Q8L7S8) At5g26740, partial (26%)
Length = 1275
Score = 29.6 bits (65), Expect = 1.8
Identities = 10/17 (58%), Positives = 10/17 (58%)
Frame = +3
Query: 6 CFNCNEEGHIGSQCKQP 22
CFNC E GH S C P
Sbjct: 816 CFNCGESGHRASDCPNP 866
>TC227589
Length = 547
Score = 29.6 bits (65), Expect = 1.8
Identities = 10/19 (52%), Positives = 11/19 (57%)
Frame = +2
Query: 6 CFNCNEEGHIGSQCKQPKR 24
CFNC E+GH C KR
Sbjct: 47 CFNCGEDGHAAVNCSAAKR 103
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 29.3 bits (64), Expect = 2.4
Identities = 9/14 (64%), Positives = 10/14 (71%)
Frame = +2
Query: 6 CFNCNEEGHIGSQC 19
CFNC EEGH +C
Sbjct: 545 CFNCGEEGHFAREC 586
>TC214039 homologue to UP|Q9M4R0 (Q9M4R0) Ubiquitin conjugating protein,
partial (67%)
Length = 639
Score = 29.3 bits (64), Expect = 2.4
Identities = 15/45 (33%), Positives = 24/45 (53%)
Frame = +3
Query: 104 VCLRCPLSMFGHDFEMDLVCLHLSGMDVILGMNWLEYNHVHINCF 148
+CL C + +F +C +SG +L M WL Y+H+ +CF
Sbjct: 93 LCLACFIQIFMLMQVFVWICYKISG---VLYMMWLLYSHLSRHCF 218
>TC233976
Length = 763
Score = 28.9 bits (63), Expect = 3.1
Identities = 8/27 (29%), Positives = 16/27 (58%)
Frame = +1
Query: 3 DIVCFNCNEEGHIGSQCKQPKRAPTTG 29
++ CFN +GH+ + C P++ +G
Sbjct: 322 EVTCFNYQGKGHLSTNCPHPRKEKRSG 402
>TC214169 similar to UP|Q8LF25 (Q8LF25) DNA-damage inducible protein
DDI1-like, partial (90%)
Length = 1604
Score = 28.9 bits (63), Expect = 3.1
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +2
Query: 52 INNTPLVAIIDTGATHCFIAFDCVSALGL 80
+N PL A +D+GA I+ C LGL
Sbjct: 764 VNGVPLKAFVDSGAQSTIISKSCAERLGL 850
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 28.9 bits (63), Expect = 3.1
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Frame = +2
Query: 6 CFNCNEEGHIGSQCK-QPKRAPT-TGRVFAMAGTQTENEDR 44
C+ C E GHI CK PK+ T GR ++ + ++ + R
Sbjct: 380 CYRCGERGHIERNCKNSPKKLSTRRGRSYSRSPVRSHSPRR 502
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.140 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,357,344
Number of Sequences: 63676
Number of extensions: 194339
Number of successful extensions: 1292
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1259
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1288
length of query: 311
length of database: 12,639,632
effective HSP length: 97
effective length of query: 214
effective length of database: 6,463,060
effective search space: 1383094840
effective search space used: 1383094840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC130807.9


