

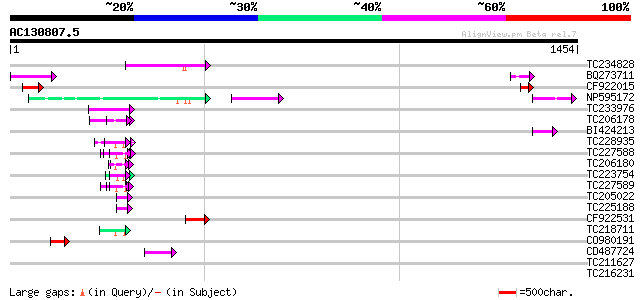
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.5 + phase: 0 /pseudo
(1454 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC234828 192 1e-48
BQ273711 95 2e-19
CF922015 80 5e-15
NP595172 polyprotein [Glycine max] 60 9e-09
TC233976 59 1e-08
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 59 2e-08
BI424213 58 3e-08
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 56 1e-07
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 54 5e-07
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 52 2e-06
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 52 2e-06
TC227589 51 3e-06
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 48 3e-05
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 48 4e-05
CF922531 45 2e-04
TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), ... 45 3e-04
CO980191 45 3e-04
CD487724 43 9e-04
TC211627 43 0.001
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 41 0.003
>TC234828
Length = 857
Score = 192 bits (487), Expect = 1e-48
Identities = 103/260 (39%), Positives = 143/260 (54%), Gaps = 43/260 (16%)
Frame = +3
Query: 297 NCNEEGHISTQCTQPKKDRTGGKVFALTGTQTTNEDRLIRGTCFFNSTPLIAIIDTGATH 356
N G++S + + +VFA++G++ D LIRG C L + D+GATH
Sbjct: 66 NSTRSGNVSNNNISGGRPKVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATH 245
Query: 357 CFIALESAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLRCPTSMFGRDFVVDLVCLPLT 416
FI+ +LGL +++ +MVV TP VTTS VCL+CP + GR F+ DL+CLPL
Sbjct: 246 SFISHACVERLGLCATELPYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLA 425
Query: 417 GMDVIFGMNWLEYNRVHINCFSKTMHF------------SSAEEE--------------- 449
+DVI GM+WL N + ++C K + F +A EE
Sbjct: 426 HLDVILGMDWLSTNHIFLDCKEKMLVFGGDVVPSEPLKEDAANEETEDVRTYMVLFSMYV 605
Query: 450 ----------------EVFPDEIPDVPPEREVEFSIDLVPGTKPMSMAPYRMSSSELAEL 493
EVFPD++ ++PPEREVEF ID+VPG P+S+APYRMS ELAE+
Sbjct: 606 EEDAEVSCIPVVSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPVELAEV 785
Query: 494 KKQLED*LDKKFVRPNVSPW 513
K Q++D L K+FVRP+ SPW
Sbjct: 786 KAQVQDLLSKQFVRPSASPW 845
>BQ273711
Length = 409
Score = 95.1 bits (235), Expect = 2e-19
Identities = 47/121 (38%), Positives = 65/121 (52%), Gaps = 3/121 (2%)
Frame = -2
Query: 1 MAGRN---DAAIAGALEAIAQAVEQQPQASNGEVRMLETFLRNHPPAFKGMYDPDGAQSW 57
MAGR + + + A+ + + Q+ E R L F +NHPP F G YDP+GA+ W
Sbjct: 363 MAGRGRDQNRDVLDRITAVLETLVQERDVEPAEYRGLMAFRKNHPPKFSGDYDPEGARLW 184
Query: 58 LKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWAVFRREFLNR 117
L E E+IF AM C E KV + T ML EA++WW + P V+ W F+ +FL
Sbjct: 183 LAETEKIFEAMGCLEEHKVLYATFMLQGEAENWWKFVKPSFVAPGGVIPWNAFKDKFLEN 4
Query: 118 Y 118
Y
Sbjct: 3 Y 1
Score = 55.1 bits (131), Expect = 2e-07
Identities = 28/61 (45%), Positives = 34/61 (54%)
Frame = -2
Query: 1285 IAAALEAIAQAVGQQPQAGNGEVRMLETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRTD 1344
I A LE + Q +P E R L F +NHPP F G YDP+GA+ WL E E+IF
Sbjct: 318 ITAVLETLVQERDVEP----AEYRGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAM 151
Query: 1345 G 1345
G
Sbjct: 150 G 148
>CF922015
Length = 172
Score = 80.5 bits (197), Expect = 5e-15
Identities = 33/54 (61%), Positives = 45/54 (83%)
Frame = -3
Query: 34 LETFLRNHPPAFKGMYDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEA 87
L+ F RN+PP FKG YDP+GA++WL+E+E+IFR M+C + +KV F THMLA+EA
Sbjct: 164 LDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFRVMECQDHQKVLFATHMLADEA 3
Score = 55.1 bits (131), Expect = 2e-07
Identities = 21/33 (63%), Positives = 29/33 (87%)
Frame = -3
Query: 1310 LETFLRNHPPAFKGMYDPDGAQSWLKEVERIFR 1342
L+ F RN+PP FKG YDP+GA++WL+E+E+IFR
Sbjct: 164 LDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFR 66
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 59.7 bits (143), Expect = 9e-09
Identities = 41/116 (35%), Positives = 58/116 (49%), Gaps = 4/116 (3%)
Frame = +1
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
++DGQSE + LE LR E W LP EF YN YH S+GM PF ALYG
Sbjct: 3715 QSDGQSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYG--- 3885
Query: 1402 RTPLCWFESGERVVLGPEIVQQTTEKVKMIQEKMKVSHSR----QKSYHDKRRKDL 1453
R P + E+ +Q T++ ++ K+K++ +R K DK+R D+
Sbjct: 3886 REPPTLTRQACSIDDPAEVREQLTDRDALL-AKLKINLTRAQQVMKRQADKKRLDV 4050
Score = 47.4 bits (111), Expect = 5e-05
Identities = 103/546 (18%), Positives = 190/546 (33%), Gaps = 81/546 (14%)
Frame = +1
Query: 49 YDPDGAQSWLKEVERIFRAMQCSEVKKVRFGTHMLAEEADDWWVSLLPVLEQDEAVVTWA 108
+D W+ + E+ F + ++ + L ++ W+ +L++ E +W
Sbjct: 313 FDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWY----QMLQKTEPFSSWQ 480
Query: 109 VFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVELAKFYPHYTAETAEFSKC 168
F R L F +L +V EY +F L +AE C
Sbjct: 481 AFTRA-LELDFGPSAYDCPRATLFKLN-QSATVNEYYMQFTALVNRVDGLSAEA--ILDC 648
Query: 169 IKFENGLRANIKRAIGYQQIRIFSDLVSRCRICEEDTKAHYKVMS----ERRGKGRQSRP 224
F +GL+ I R + + R + V+ ++ EE + K + R S
Sbjct: 649 --FVSGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPKTKTFSNLARNFTSNTSAT 822
Query: 225 KPYSAPADKGKQRLNDERRPNRRDA--SAEIVCFKCGEKGHK----SNVCTKDEKK-CFR 277
+ Y K ND +PN + F + K + + + EK C+
Sbjct: 823 QKYPPTNQK-----NDNPKPNLPPLLPTPSTKPFNLRNQNIKKISPAEIQLRREKNLCYF 987
Query: 278 CGQKGHLLADCKRGDVVCYNCNEEGHISTQ----CTQPKKDRTGGKVFALTGTQTTNEDR 333
C +K C V+ E T T+ +L + +N
Sbjct: 988 CDEKFSPAHKCPNRQVMLLQLEETDEDQTDEQVMVTEEANMDDDTHHLSLNAMRGSNGVG 1167
Query: 334 LIRGTCFFNSTPLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMVVETPAKGSVTTSL- 392
IR T + ++D G++ FI A L L V V+ G + ++
Sbjct: 1168TIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAPNLRVLV--GNGQILSAEG 1341
Query: 393 VCLRCPTSMFGRDFVVDLVCLPLTGMDVIFGMNWL------------------------- 427
+ + P + G++ V + L ++G DVI G WL
Sbjct: 1342IVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADYAALTLKFFQNDKFIT 1521
Query: 428 -------EYNRVHINCFSKTMHFSSAEE--------EEVFPDEIPDVP------------ 460
E + ++ F + + S EE +EV D + D+P
Sbjct: 1522LQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKEVPEDTLKDLPTNIDPELAILLH 1701
Query: 461 -------------PEREVEFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVR 507
P+RE + +I L G+ P+ + PYR ++ +++K +++ L + ++
Sbjct: 1702TYAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQ 1881
Query: 508 PNVSPW 513
P+ SP+
Sbjct: 1882PSNSPF 1899
Score = 46.2 bits (108), Expect = 1e-04
Identities = 46/134 (34%), Positives = 68/134 (50%)
Frame = +2
Query: 568 *GLVTIKLR*RMKICRRRPLGHVMVITNTR*CLSVLLMRLVCSWSI*TEFFMPFWISLWV 627
*G TIK ++I R+ L H+M I N *C VLLM S + *T++F +L++
Sbjct: 2063 *GQDTIKFWYSLRIERKLLLEHIMAIMNG**CHLVLLMHQPHSNA**TKYFSLLSENLFL 2242
Query: 628 YSLMIS*FILRLKRSMQNI*GLYCKY*KRKSCMLSC*NVNSG*VK*VFLATLFLVVVLLL 687
LM S*+I+ L R + + L + * S +L C NV+ K + AT +L
Sbjct: 2243 CFLMTS*YIVLLGRIISSTWNLCYRP*SNISYLLDCPNVHLEIQKLITWATRYLG*EFPW 2422
Query: 688 TLQKLMQYHNGRLR 701
+Q+ QY G L+
Sbjct: 2423 RIQRYKQY*IGLLQ 2464
>TC233976
Length = 763
Score = 59.3 bits (142), Expect = 1e-08
Identities = 34/128 (26%), Positives = 56/128 (43%), Gaps = 9/128 (7%)
Frame = +1
Query: 202 EEDTKAHYKVMSERRGKGRQ----SRPKPYSAPADK-----GKQRLNDERRPNRRDASAE 252
+++ A YK + GK ++ SR KPYSAP + G++
Sbjct: 25 QQEKVAFYKNANASHGKEKKPMTHSRAKPYSAPLEYENHYGGQRTSGGHHLAGGSSQLVN 204
Query: 253 IVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPK 312
V G G + +C +CG+ GH +C +V C+N +GH+ST C P+
Sbjct: 205 RVSQPAGRGGSGAPAIVTTPLRCRKCGRLGHNAHECTYREVTCFNYQGKGHLSTNCPHPR 384
Query: 313 KDRTGGKV 320
K++ G +
Sbjct: 385 KEKRSGSL 408
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 58.9 bits (141), Expect = 2e-08
Identities = 34/118 (28%), Positives = 54/118 (44%), Gaps = 2/118 (1%)
Frame = +2
Query: 204 DTKAHYKVMSERRGKGRQSRPKPYSAP--ADKGKQRLNDERRPNRRDASAEIVCFKCGEK 261
+ K +K+ S+ R + R P +D+ R RR +RR S + +C C
Sbjct: 185 ERKKKWKMSSDSRSRSRSRSRSPMDRKIRSDRFSYRDAPYRRDSRRGFSRDNLCKNCKRP 364
Query: 262 GHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKKDRTGGK 319
GH + C + C CG GH+ ++C + C+NC E GH+++ C T GK
Sbjct: 365 GHYARECP-NVAICHNCGLPGHIASECTTKSL-CWNCKEPGHMASSCPNEGICHTCGK 532
Score = 52.4 bits (124), Expect = 1e-06
Identities = 24/71 (33%), Positives = 40/71 (55%), Gaps = 6/71 (8%)
Frame = +2
Query: 248 DASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKR-----GDV-VCYNCNEE 301
+ + + +C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC ++
Sbjct: 437 ECTTKSLCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQ 613
Query: 302 GHISTQCTQPK 312
GHI+ +CT K
Sbjct: 614 GHIAAECTNEK 646
Score = 40.8 bits (94), Expect = 0.004
Identities = 21/87 (24%), Positives = 32/87 (36%), Gaps = 25/87 (28%)
Frame = +2
Query: 247 RDASAEIVCFKCGEKGHKSNVCTKD-------------------------EKKCFRCGQK 281
RD + +C C GH + C K + C C Q
Sbjct: 683 RDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGGGGGYRDVVCRNCQQL 862
Query: 282 GHLLADCKRGDVVCYNCNEEGHISTQC 308
GH+ DC ++C+NC GH++ +C
Sbjct: 863 GHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 38.9 bits (89), Expect = 0.017
Identities = 28/102 (27%), Positives = 36/102 (34%), Gaps = 44/102 (43%)
Frame = +2
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCK------------------------ 289
+C C ++GH + CT +EK C C + GHL DC
Sbjct: 590 LCNNCYKQGHIAAECT-NEKACNNCRKTGHLARDCPNDPICNLCNVSGHVARQCPKANVL 766
Query: 290 -------------RG-------DVVCYNCNEEGHISTQCTQP 311
RG DVVC NC + GH+S C P
Sbjct: 767 GDRSGGGGGGGGARGGGGGGYRDVVCRNCQQLGHMSRDCMGP 892
Score = 38.1 bits (87), Expect = 0.028
Identities = 15/40 (37%), Positives = 20/40 (49%)
Frame = +2
Query: 252 EIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRG 291
++VC C + GH S C C CG +GHL +C G
Sbjct: 833 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 952
>BI424213
Length = 426
Score = 57.8 bits (138), Expect = 3e-08
Identities = 27/64 (42%), Positives = 38/64 (59%)
Frame = +1
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFEALYGQRC 1401
+TDGQ++ +SL LLR + +WD +LP +EF YN G H + +PFE +YG
Sbjct: 73 QTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEFAYNRGVHRTTKQSPFEVVYGFNP 252
Query: 1402 RTPL 1405
TPL
Sbjct: 253 LTPL 264
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 55.8 bits (133), Expect = 1e-07
Identities = 25/80 (31%), Positives = 38/80 (47%), Gaps = 13/80 (16%)
Frame = +1
Query: 244 PNRRDASAEIVCFKCGEKGHKSNVC------TKDEKKCFRCGQKGHLLADC----KRGDV 293
P + + +C +C +GH++ C KD K C+ CG+ GH L C + G
Sbjct: 313 PEKAEWEKNKICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQEGGT 492
Query: 294 V---CYNCNEEGHISTQCTQ 310
C+ CN+ GH+S C Q
Sbjct: 493 KFAECFVCNQRGHLSKNCPQ 552
Score = 51.2 bits (121), Expect = 3e-06
Identities = 34/116 (29%), Positives = 50/116 (42%), Gaps = 11/116 (9%)
Frame = +1
Query: 218 KGRQSRPKPYSAPADKGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKD-----E 272
K ++ PKP G ++ + R P + + CF C H + +C +
Sbjct: 190 KRKRPEPKP-------GSRKRHLLRVPGMKPGES---CFICKAMDHIAKLCPEKAEWEKN 339
Query: 273 KKCFRCGQKGHLLADCK------RGDVVCYNCNEEGHISTQCTQPKKDRTGGKVFA 322
K C RC ++GH +C + CYNC E GH TQC P ++ GG FA
Sbjct: 340 KICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQE--GGTKFA 501
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 53.9 bits (128), Expect = 5e-07
Identities = 28/97 (28%), Positives = 42/97 (42%), Gaps = 11/97 (11%)
Frame = +1
Query: 233 KGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKD-------EKKCFRCGQKGHLL 285
KG R D + + + +C KCG GH C D E +C+ C + GHL
Sbjct: 199 KGGHRAKDCPEKHTSTSKSIAICLKCGNSGHDIFSCRNDYSQDDLKEIQCYVCKRLGHLC 378
Query: 286 A----DCKRGDVVCYNCNEEGHISTQCTQPKKDRTGG 318
D G++ CY C + GH C++ + + T G
Sbjct: 379 CVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITSG 489
Score = 50.4 bits (119), Expect = 6e-06
Identities = 30/85 (35%), Positives = 38/85 (44%), Gaps = 13/85 (15%)
Frame = +1
Query: 242 RRPNRRDASAEIVCFKCGEKGHKSNVCTKD----EKKCFRCGQKGHLLADCKR-GDVV-- 294
R +D EI C+ C GH V T D E C++CGQ GH C R D +
Sbjct: 307 RNDYSQDDLKEIQCYVCKRLGHLCCVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITS 486
Query: 295 ------CYNCNEEGHISTQCTQPKK 313
C+ C EEGH + +CT K
Sbjct: 487 GATPSSCFKCGEEGHFARECTSSIK 561
Score = 48.1 bits (113), Expect = 3e-05
Identities = 25/71 (35%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Frame = +1
Query: 255 CFKCGEKGHKSNVCT--KDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPK 312
CF CGE+GH + C+ K +K C+ CG GH C + C+ C + GH + C P+
Sbjct: 61 CFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQCSKVQ-DCFICKKGGHRAKDC--PE 231
Query: 313 KDRTGGKVFAL 323
K + K A+
Sbjct: 232 KHTSTSKSIAI 264
Score = 32.7 bits (73), Expect = 1.2
Identities = 18/47 (38%), Positives = 22/47 (46%)
Frame = +1
Query: 237 RLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGH 283
RL DE +S CFKCGE+GH + CT K R + H
Sbjct: 463 RLRDEITSGATPSS----CFKCGEEGHFARECTSSIKSGKRNWESSH 591
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 51.6 bits (122), Expect = 2e-06
Identities = 26/64 (40%), Positives = 36/64 (55%)
Frame = +3
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
+C C ++GH + CT +EK C C + GHL DC D +C CN GH++ QC PK
Sbjct: 75 LCNNCYKQGHIAAECT-NEKACNNCRKTGHLARDCP-NDPICNLCNVSGHVARQC--PKA 242
Query: 314 DRTG 317
+ G
Sbjct: 243 NVLG 254
Score = 44.3 bits (103), Expect = 4e-04
Identities = 18/57 (31%), Positives = 31/57 (53%), Gaps = 6/57 (10%)
Frame = +3
Query: 258 CGEKGHKSNVCTK------DEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQC 308
CG+ GH++ C+ D + C C ++GH+ A+C + C NC + GH++ C
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC-TNEKACNNCRKTGHLARDC 176
Score = 42.0 bits (97), Expect = 0.002
Identities = 21/84 (25%), Positives = 32/84 (38%), Gaps = 22/84 (26%)
Frame = +3
Query: 247 RDASAEIVCFKCGEKGHKSNVCTKD----------------------EKKCFRCGQKGHL 284
RD + +C C GH + C K + C C Q GH+
Sbjct: 168 RDCPNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGGYRDVVCRNCQQLGHM 347
Query: 285 LADCKRGDVVCYNCNEEGHISTQC 308
DC ++C+NC GH++ +C
Sbjct: 348 SRDCMGPLMICHNCGGRGHLAYEC 419
Score = 38.1 bits (87), Expect = 0.028
Identities = 15/40 (37%), Positives = 20/40 (49%)
Frame = +3
Query: 252 EIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHLLADCKRG 291
++VC C + GH S C C CG +GHL +C G
Sbjct: 309 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 428
Score = 36.6 bits (83), Expect = 0.083
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 6/41 (14%)
Frame = +3
Query: 278 CGQKGHLLADCKR-----GDV-VCYNCNEEGHISTQCTQPK 312
CG+ GH +C GD+ +C NC ++GHI+ +CT K
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEK 131
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 51.6 bits (122), Expect = 2e-06
Identities = 28/95 (29%), Positives = 37/95 (38%), Gaps = 22/95 (23%)
Frame = +2
Query: 246 RRDASAEIVCFKCGEKGHKSNVCTKDEKK----------CFRCGQKGHLLADCKR----- 290
RR+ C++CGE GH + C + CF CG GHL DC R
Sbjct: 17 RRNGGGGAACYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCVRGGGGS 196
Query: 291 -------GDVVCYNCNEEGHISTQCTQPKKDRTGG 318
G C+ C GH++ C K + GG
Sbjct: 197 VGIGGGGGGGSCFRCGGFGHMARDCATGKGNIGGG 301
Score = 50.1 bits (118), Expect = 7e-06
Identities = 24/80 (30%), Positives = 33/80 (41%), Gaps = 24/80 (30%)
Frame = +2
Query: 255 CFKCGEKGHKSNVCTKDEKK---------CFRCGQKGHLLADCKR--------------- 290
CF+CG GH + C + CFRCG+ GHL DC
Sbjct: 230 CFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDCGMEGGRFGGGGGSGGGG 409
Query: 291 GDVVCYNCNEEGHISTQCTQ 310
G C+NC + GH + +C +
Sbjct: 410 GKSTCFNCGKPGHFARECVE 469
>TC227589
Length = 547
Score = 51.2 bits (121), Expect = 3e-06
Identities = 26/89 (29%), Positives = 38/89 (42%), Gaps = 11/89 (12%)
Frame = +2
Query: 233 KGKQRLNDERRPNRRDASAEIVCFKCGEKGHKSNVCTKD-------EKKCFRCGQKGHLL 285
KG R D + + + +C KCG GH C D E +C+ C + GHL
Sbjct: 185 KGGHRAKDCLEKHTSRSKSVAICLKCGNSGHDMFSCRNDYSPDDLKEIQCYVCKRVGHLC 364
Query: 286 A----DCKRGDVVCYNCNEEGHISTQCTQ 310
D G++ CY C + GH C++
Sbjct: 365 CVNTDDATPGEISCYKCGQLGHTGLACSK 451
Score = 47.8 bits (112), Expect = 4e-05
Identities = 29/77 (37%), Positives = 35/77 (44%), Gaps = 13/77 (16%)
Frame = +2
Query: 248 DASAEIVCFKCGEKGHKSNVCTKD----EKKCFRCGQKGHL-LADCKRGDVV-------- 294
D EI C+ C GH V T D E C++CGQ GH LA K D +
Sbjct: 311 DDLKEIQCYVCKRVGHLCCVNTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSS 490
Query: 295 CYNCNEEGHISTQCTQP 311
C C E GH + +CT P
Sbjct: 491 CCKCGEAGHFAQECTSP 541
Score = 47.8 bits (112), Expect = 4e-05
Identities = 22/64 (34%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Frame = +2
Query: 255 CFKCGEKGHKSNVCT--KDEKKCFRCGQKGHLLADCKRGDVVCYNCNEEGHISTQCTQPK 312
CF CGE GH + C+ K +K C+ CG GH C + C+ C + GH + C +
Sbjct: 47 CFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQCTKAQ-DCFICKKGGHRAKDCLEKH 223
Query: 313 KDRT 316
R+
Sbjct: 224 TSRS 235
Score = 40.8 bits (94), Expect = 0.004
Identities = 21/54 (38%), Positives = 28/54 (50%), Gaps = 10/54 (18%)
Frame = +2
Query: 245 NRRDAS-AEIVCFKCGEKGHKSNVCTK--DE-------KKCFRCGQKGHLLADC 288
N DA+ EI C+KCG+ GH C+K DE C +CG+ GH +C
Sbjct: 371 NTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSSCCKCGEAGHFAQEC 532
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 48.1 bits (113), Expect = 3e-05
Identities = 22/43 (51%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = +2
Query: 274 KCFRCGQKGHLLADCKRGD--VVCYNCNEEGHISTQC-TQPKK 313
+CF CG GH DCK GD CY C E GHI C PKK
Sbjct: 311 RCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCKNSPKK 439
Score = 42.4 bits (98), Expect = 0.002
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 2/37 (5%)
Frame = +2
Query: 255 CFKCGEKGHKSNVCTKDE--KKCFRCGQKGHLLADCK 289
CF CG GH + C + KC+RCG++GH+ +CK
Sbjct: 314 CFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCK 424
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 47.8 bits (112), Expect = 4e-05
Identities = 22/43 (51%), Positives = 23/43 (53%), Gaps = 3/43 (6%)
Frame = +1
Query: 274 KCFRCGQKGHLLADCKRGD--VVCYNCNEEGHISTQC-TQPKK 313
+CF CG GH DCK GD CY C E GHI C PKK
Sbjct: 361 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKK 489
Score = 42.4 bits (98), Expect = 0.002
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 2/37 (5%)
Frame = +1
Query: 255 CFKCGEKGHKSNVCTKDE--KKCFRCGQKGHLLADCK 289
CF CG GH + C + KC+RCG++GH+ +CK
Sbjct: 364 CFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 474
>CF922531
Length = 602
Score = 45.1 bits (105), Expect = 2e-04
Identities = 23/63 (36%), Positives = 39/63 (61%), Gaps = 1/63 (1%)
Frame = -2
Query: 450 EVFPDEIP-DVPPEREVEFSIDLVPGTKPMSMAPYRMSSSELAELKKQLED*LDKKFVRP 508
++FP EIP +PP R +E IDLVP + YR + E E++ Q+++ L+K +V+
Sbjct: 229 DIFPKEIPLGLPPLRGIEHQIDLVPRASLPNRPTYRTNPQETKEIESQVKELLEKGWVQE 50
Query: 509 NVS 511
++S
Sbjct: 49 SLS 41
>TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), partial (6%)
Length = 1204
Score = 44.7 bits (104), Expect = 3e-04
Identities = 28/97 (28%), Positives = 38/97 (38%), Gaps = 16/97 (16%)
Frame = -3
Query: 230 PADKGKQRLNDERR----PNRRDASAEIVCFKCGEKGHKSNVCTK------DEKKCFRCG 279
PA K ++ D R P + + + C C + GH+ C + DE+ CF CG
Sbjct: 608 PAGKTLVQITDTLRVTKSPFKHHGESSLRCRACRQPGHRFQQCQRLKCLSMDEEVCFFCG 429
Query: 280 QKGHLLADCKRGD------VVCYNCNEEGHISTQCTQ 310
+ GH L C C C GH S C Q
Sbjct: 428 EIGHSLGKCDVSQAGGGRFAKCLLCYGHGHFSYNCPQ 318
>CO980191
Length = 802
Score = 44.7 bits (104), Expect = 3e-04
Identities = 23/50 (46%), Positives = 31/50 (62%)
Frame = +1
Query: 104 VVTWAVFRREFLNRYFLEDV*GRKEIEFLELK*GDISVVEYAAKFVELAK 153
V+T VF+ FL ++ ED+* K+ EFLELK G++ V Y F EL K
Sbjct: 361 VIT*EVFKEIFLEKHLHEDI*NHKKTEFLELKHGNMIVANYLVNFDELPK 510
>CD487724
Length = 676
Score = 43.1 bits (100), Expect = 9e-04
Identities = 24/84 (28%), Positives = 42/84 (49%)
Frame = +3
Query: 345 PLIAIIDTGATHCFIALESAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLRCPTSMFGR 404
PL+ ++D G+TH F+ +LGL V+ T+ +C P S+
Sbjct: 21 PLVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTT-ICEAIPISIQNI 197
Query: 405 DFVVDLVCLPLTGMDVIFGMNWLE 428
+F+V L LP+ G +++ G+ WL+
Sbjct: 198 EFLVHLYVLPIVGANIVLGVQWLK 269
>TC211627
Length = 1034
Score = 42.7 bits (99), Expect = 0.001
Identities = 23/53 (43%), Positives = 28/53 (52%)
Frame = +2
Query: 1342 RTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNGYHSSIGMAPFE 1394
+TDGQ+E + LE LR V + W L L E YN HS IG +PFE
Sbjct: 80 QTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLAE*CYNTSVHSGIGFSPFE 238
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 41.2 bits (95), Expect = 0.003
Identities = 16/47 (34%), Positives = 22/47 (46%), Gaps = 13/47 (27%)
Frame = +3
Query: 275 CFRCGQKGHLLADCKR-------------GDVVCYNCNEEGHISTQC 308
C++CG+ GH+ DC + G CYNC E GH + C
Sbjct: 231 CYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDC 371
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.351 0.154 0.543
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,654,489
Number of Sequences: 63676
Number of extensions: 1098405
Number of successful extensions: 9210
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 6441
Number of HSP's successfully gapped in prelim test: 243
Number of HSP's that attempted gapping in prelim test: 2197
Number of HSP's gapped (non-prelim): 7294
length of query: 1454
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1345
effective length of database: 5,698,948
effective search space: 7665085060
effective search space used: 7665085060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC130807.5


