

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.4 - phase: 0
(329 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
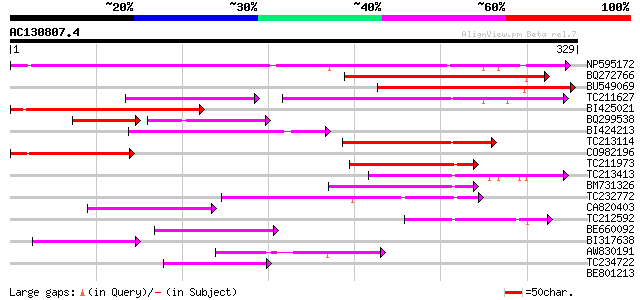
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 203 8e-53
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 159 2e-39
BU549069 159 2e-39
TC211627 78 5e-29
BI425021 100 7e-22
BQ299538 51 7e-16
BI424213 81 7e-16
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 69 3e-12
CO982196 67 8e-12
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 67 1e-11
TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 63 2e-10
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 55 6e-08
TC232772 weakly similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial ... 54 7e-08
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 54 1e-07
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 53 2e-07
BE660092 weakly similar to GP|9884624|dbj retroelement pol polyp... 52 4e-07
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 52 5e-07
AW830191 47 2e-05
TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (... 44 1e-04
BE801213 weakly similar to GP|6691193|gb| F7F22.17 {Arabidopsis ... 32 0.004
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 203 bits (517), Expect = 8e-53
Identities = 127/348 (36%), Positives = 186/348 (52%), Gaps = 23/348 (6%)
Frame = +1
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF+ LPN+ G I VV+DRL K AHFIP+ Y +AE ++ +IVKLHG+P SI
Sbjct: 3442 MDFITGLPNSF-GLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRSI 3618
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAW 120
VSDRD FTS FW+ L G+ L +SSAYHPQ+DGQS+ + LE LR E W
Sbjct: 3619 VSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHPKGW 3798
Query: 121 DSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVK 180
LP EF YN +YH S+GM PF+ALYGR P S+ ++ + T++
Sbjct: 3799 VKALPWAEFWYNTAYHMSLGMTPFRALYGRE---PPTLTRQACSIDDPAEVREQLTDRDA 3969
Query: 181 MIRK---KMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGHAL-KSRKLTPKFI 236
++ K + +Q K DK+R + FQ GD V +++ P L K++KL+ ++
Sbjct: 3970 LLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRYF 4149
Query: 237 GPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKY---VADPSHVIP----------- 282
GP+++ ++G VAY++ L P + +H VFHVSQL+ + DP +P
Sbjct: 4150 GPFKVLAKIGDVAYKLEL-PSAARIHPVFHVSQLKPFNGTAQDPYLPLPLTVTEMGPVMQ 4326
Query: 283 -----RDDVQVRDNLTVETLPLVKVVWDGATGESLTWELESKMRDSYP 325
+ +R + +E + V W+ + TWE ++ SYP
Sbjct: 4327 PVKILASRIIIRGHNQIEQ---ILVQWENGLQDEATWEDIEDIKASYP 4461
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo},
partial (9%)
Length = 410
Score = 159 bits (402), Expect = 2e-39
Identities = 81/136 (59%), Positives = 98/136 (71%), Gaps = 17/136 (12%)
Frame = -3
Query: 195 SYHDKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGL 254
SYHDKRRK LEF+ GDHVFLRVTP TGVG ALKS KLTP FIGP+QI ++V VAY++ L
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 255 PPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLP---------------- 298
PP L++LH+VFHVSQ+ KY+ DPSH++ D VQV++NL ETLP
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 299 -LVKVVWDGATGESLT 313
LVKV+W GA+GE T
Sbjct: 48 LLVKVIWGGASGEEAT 1
>BU549069
Length = 615
Score = 159 bits (401), Expect = 2e-39
Identities = 80/132 (60%), Positives = 94/132 (70%), Gaps = 17/132 (12%)
Frame = -1
Query: 214 LRVTPMTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKY 273
L+VTP TGVG ALKSRKLTP FIG +QI +R VAY++ LPP LSNLH+VFHVSQLR Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 274 VADPSHVIPRDDVQVRDNLTVETL-----------------PLVKVVWDGATGESLTWEL 316
+ DPSHV+ DDVQV++NLT ETL PLVKV+W G +GE TWEL
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 317 ESKMRDSYPKLF 328
ES+MR +YP LF
Sbjct: 255 ESQMRVAYPSLF 220
>TC211627
Length = 1034
Score = 77.8 bits (190), Expect(2) = 5e-29
Identities = 56/183 (30%), Positives = 80/183 (43%), Gaps = 17/183 (9%)
Frame = +3
Query: 159 FESGESVVLGPD-LVHETTEKVKMIRKKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVT 217
+ +G S V D ++H + +++ Q K D R+ L F GD V++R+
Sbjct: 276 YSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL* 455
Query: 218 PMTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKY---- 273
P KL+ +F GPYQI RVG VAYR+ LPP S +H +FHVS L+ +
Sbjct: 456 PYRQTSIQSTYTKLSKRFYGPYQIQARVGQVAYRLQLPP-TSKIHPIFHVSLLKVHHGPI 632
Query: 274 -----VADPSHVIPRDDVQ-------VRDNLTVETLPLVKVVWDGATGESLTWELESKMR 321
P VQ D T +P V V W E TWE ++++
Sbjct: 633 PPELLALPPFSTTNHPLVQPLQFLDWKMDESTTPPIPQVLVQWTNLAPEDTTWESWTQLK 812
Query: 322 DSY 324
D Y
Sbjct: 813 DIY 821
Score = 67.8 bits (164), Expect(2) = 5e-29
Identities = 36/78 (46%), Positives = 44/78 (56%)
Frame = +2
Query: 68 FTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLI 127
F S W L G+KLR S+AYHPQTDGQ++ + LE LR V + W L L
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 128 EFTYNNSYHSSIGMAPFK 145
E YN S HS IG +PF+
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
>BI425021
Length = 426
Score = 100 bits (250), Expect = 7e-22
Identities = 51/113 (45%), Positives = 72/113 (63%)
Frame = -1
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF++ LP GH +I+VVV+R K H + + +A ++++ ++KLHG P SI
Sbjct: 381 MDFIVGLP-PYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLFLNIVIKLHGFPRSI 205
Query: 61 VSDRDPRFTSRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCV 113
VSDRDP F S FW+ L G+ LR+SSAYHPQTDGQ++ + +E LR V
Sbjct: 204 VSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIEQYLRAFV 46
>BQ299538
Length = 426
Score = 50.8 bits (120), Expect(2) = 7e-16
Identities = 22/40 (55%), Positives = 27/40 (67%)
Frame = +1
Query: 37 YPVAQLAEIYIHNIVKLHGVPSSIVSDRDPRFTSRFWKSL 76
Y LAEI+ +V LHGVP+S++SD DP F S FWK L
Sbjct: 13 YSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
Score = 50.4 bits (119), Expect(2) = 7e-16
Identities = 26/71 (36%), Positives = 41/71 (57%)
Frame = +3
Query: 81 GSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIG 140
G+ L++S++YHP DGQ+ LE LR V ++ L E+ YN ++H+S G
Sbjct: 144 GTYLKMSTSYHP*IDGQTVN--HCLETFLRCFVADQPKM*VQWLSWAEYWYNTNFHASTG 317
Query: 141 MAPFKALYGRR 151
PF+ +YGR+
Sbjct: 318 TTPFEVVYGRK 350
>BI424213
Length = 426
Score = 80.9 bits (198), Expect = 7e-16
Identities = 45/117 (38%), Positives = 61/117 (51%)
Frame = +1
Query: 70 SRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEF 129
S FWK+L LG+KL S+ HPQTDGQ+K +SL LLR + +WD +LP +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 130 TYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVKMIRKKM 186
YN H + +PF+ +YG TPL + L +H+ E KKM
Sbjct: 184 AYNRGVHRTTKQSPFEVVYGFNPLTPLDLI----PLPLDTSFIHKEGESRSEFVKKM 342
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 68.9 bits (167), Expect = 3e-12
Identities = 37/90 (41%), Positives = 56/90 (62%), Gaps = 1/90 (1%)
Frame = +3
Query: 194 KSYHDKRRKALEFQEGDHVFLRVTPMTGVGHALK-SRKLTPKFIGPYQISERVGTVAYRV 252
K+Y D+ R+A+ GD V+L++ P A K + KL+P+F GPYQI +++G VA+ +
Sbjct: 6 KAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFEL 185
Query: 253 GLPPHLSNLHDVFHVSQLRKYVADPSHVIP 282
LPP +H VFH S L+K VA ++ P
Sbjct: 186 DLPP-ARKIHPVFHASLLKKAVAATANPQP 272
>CO982196
Length = 812
Score = 67.4 bits (163), Expect = 8e-12
Identities = 35/72 (48%), Positives = 47/72 (64%)
Frame = +1
Query: 1 MDFVMSLPNTTRGHDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSI 60
MDF+ LP +G D+I VVVDRL K AHF ++ Y ++AE++I +V+LHG P+SI
Sbjct: 598 MDFIGGLPKA-QGKDNILVVVDRLTKYAHFFALSHPYTAKEVAELFIKELVRLHGFPASI 774
Query: 61 VSDRDPRFTSRF 72
VSD F S F
Sbjct: 775 VSDXXRLFMSLF 810
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 66.6 bits (161), Expect = 1e-11
Identities = 33/76 (43%), Positives = 52/76 (68%), Gaps = 1/76 (1%)
Frame = +1
Query: 198 DKRRKALEFQEGDHVFLRVTPMTGVGHALK-SRKLTPKFIGPYQISERVGTVAYRVGLPP 256
+KRR+ +E+ GD VFL++ P A + + KL+P+F P+Q+ +VGT+AY++ LP
Sbjct: 112 NKRRRDIEYVVGD*VFLKMQPYRRRSLAKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPS 291
Query: 257 HLSNLHDVFHVSQLRK 272
H+ +H VFHVS L+K
Sbjct: 292 HI-KIHPVFHVSLLKK 336
>TC213413 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (5%)
Length = 761
Score = 63.2 bits (152), Expect = 2e-10
Identities = 44/133 (33%), Positives = 67/133 (50%), Gaps = 17/133 (12%)
Frame = +1
Query: 209 GDHVFLRVTP-MTGVGHALKSRKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHV 267
GD V +++ P G KLT ++ GP+++ ER+G V YR+ L H S +H VFHV
Sbjct: 7 GDWVLVKLRPHRQGSASETTYSKLTKRYYGPFEVQERLGKVVYRLKLTAH-SRIHPVFHV 183
Query: 268 SQLRKYVADP--SHVIP----RDDVQVRDNLTV---ETLP-------LVKVVWDGATGES 311
S L+ +V DP +H P R + LTV + +P +V V W A+ +
Sbjct: 184 SLLKAFVGDPETTHAGPLPVMRTEEATNTPLTVIDSKLVPADNGPRRMVLVQWPSASRQD 363
Query: 312 LTWELESKMRDSY 324
+WE +R+ Y
Sbjct: 364 ASWEDWQVLRERY 402
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 54.7 bits (130), Expect = 6e-08
Identities = 30/88 (34%), Positives = 50/88 (56%), Gaps = 1/88 (1%)
Frame = +1
Query: 186 MKASQSRQKSYHDKRRKALEFQEGDHVFLRVTP-MTGVGHALKSRKLTPKFIGPYQISER 244
++ +QS + + R+ + GD V+L++ P G KLT +F GPY + +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 245 VGTVAYRVGLPPHLSNLHDVFHVSQLRK 272
VG VA+++ LP + +H VFHVSQL++
Sbjct: 205 VGAVAFQLQLPSE-ARIHPVFHVSQLKR 285
>TC232772 weakly similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (7%)
Length = 729
Score = 54.3 bits (129), Expect = 7e-08
Identities = 38/156 (24%), Positives = 70/156 (44%), Gaps = 4/156 (2%)
Frame = +1
Query: 124 LPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKVKMIR 183
LP +EF YN + HS+ PF+ +Y TPL H E +K +
Sbjct: 7 LPHVEFAYNRAVHSTTQHFPFEVVYDFNPLTPLDSLPLSNISGFKHKDAHAKVEYIKRLH 186
Query: 184 KKMKASQSRQKSYH----DKRRKALEFQEGDHVFLRVTPMTGVGHALKSRKLTPKFIGPY 239
++ K +++ + +K RK + + D V++ + + KL P+ GP+
Sbjct: 187 EQAKTQIAKKNESYVKQTNKNRKKVVLEPSDWVWVHMRKERFPKQRMS--KLQPRGDGPF 360
Query: 240 QISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVA 275
Q+ ER+ AY++ +P + F+++ L +VA
Sbjct: 361 QVLERINYNAYKIDIPGEY-EVSSSFNIADLTPFVA 465
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 53.9 bits (128), Expect = 1e-07
Identities = 32/76 (42%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Frame = -3
Query: 46 YIHNIVKLHGVPSSIVSDRDPRFT-SRFWKSLQDALGSKLRLSSAYHPQTDGQSKRTIQS 104
+I VKLHG SSIVSD D F S FW L G+KL+ S AYHPQ D +K +
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 105 LEYLLRVCVLEEGGAW 120
+E L+ + W
Sbjct: 155 IEMNLQCLTTSKRKQW 108
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 52.8 bits (125), Expect = 2e-07
Identities = 35/100 (35%), Positives = 49/100 (49%), Gaps = 14/100 (14%)
Frame = +2
Query: 230 KLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVR 289
KL+ +F GP+Q+ ERVG VAYR+ LP + LH VFH S L+ + +P +
Sbjct: 74 KLSKRFFGPFQVVERVGKVAYRLQLPVD-AKLHPVFHCSLLKPFQGNPPDTAAPLPPTLF 250
Query: 290 DNLTVETLPL--------------VKVVWDGATGESLTWE 315
D+ +V PL V V W G + + TWE
Sbjct: 251 DHQSV-IAPLVILATRTVNDDDIEVLVQWQGLSPDDATWE 367
>BE660092 weakly similar to GP|9884624|dbj retroelement pol polyprotein-like
{Arabidopsis thaliana}, partial (13%)
Length = 378
Score = 52.0 bits (123), Expect = 4e-07
Identities = 21/72 (29%), Positives = 43/72 (59%)
Frame = -3
Query: 85 RLSSAYHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIGMAPF 144
R+S+ YHPQT+GQ++ + + ++ +L V W + L + + +Y + IGM+P+
Sbjct: 340 RVSTPYHPQTNGQAEISNREIKRILEKIVQPSRKDWSTRLDDALWAHRTAYKAPIGMSPY 161
Query: 145 KALYGRRCRTPL 156
+ ++G+ C P+
Sbjct: 160 RVVFGKACHLPV 125
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 51.6 bits (122), Expect = 5e-07
Identities = 25/63 (39%), Positives = 36/63 (56%)
Frame = -2
Query: 14 HDSIWVVVDRLRKSAHFIPINIKYPVAQLAEIYIHNIVKLHGVPSSIVSDRDPRFTSRFW 73
H +I VVVD K H + + +A ++I ++ KLHG+P S+VSD D F S FW
Sbjct: 194 HTAILVVVDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFW 15
Query: 74 KSL 76
+ L
Sbjct: 14 QEL 6
>AW830191
Length = 372
Score = 46.6 bits (109), Expect = 2e-05
Identities = 30/109 (27%), Positives = 49/109 (44%), Gaps = 10/109 (9%)
Frame = +3
Query: 120 WDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPLCWFESGESVVLGPDLVHETTEKV 179
W L EF +N +Y++S+ + PFK LYG C P +L ++ T E+V
Sbjct: 39 WPKRLSWAEFWFNTNYNNSLKLTPFKVLYG--CDPP---------HLLKGAIISSTAEEV 185
Query: 180 KMIR----------KKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTP 218
++ K A Q Y D+ R+++ GD V+L++ P
Sbjct: 186 NVMTNDRDQMLHDLKGNLAKAQNQMKYADRSRRSIPLNVGDWVYLKLQP 332
>TC234722 similar to UP|Q6WAY9 (Q6WAY9) Pol (Fragment), partial (32%)
Length = 482
Score = 43.9 bits (102), Expect = 1e-04
Identities = 19/63 (30%), Positives = 37/63 (58%)
Frame = -3
Query: 90 YHPQTDGQSKRTIQSLEYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIGMAPFKALYG 149
YHPQT+GQ++ + + ++ +L V+ W L + Y ++ + IG++PF+ +YG
Sbjct: 234 YHPQTNGQAEVSNKEIKRVLENIVVSSRKDWALKLDDAFWAYRIAFKTPIGLSPFQLVYG 55
Query: 150 RRC 152
+ C
Sbjct: 54 KAC 46
>BE801213 weakly similar to GP|6691193|gb| F7F22.17 {Arabidopsis thaliana},
partial (3%)
Length = 416
Score = 31.6 bits (70), Expect(2) = 0.004
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Frame = +3
Query: 49 NIVKLHGVPSSIVSDRDPRFTSRFWKSLQDAL---GSKLRLSSAYHPQTDGQSK 99
NI G+P ++SD F ++ L L + ++ ++YHPQT+GQ+K
Sbjct: 39 NIFSRFGMPRILISDGGSHF---YYSQLNKVLKHDSVRHKVETSYHPQTNGQAK 191
Score = 26.2 bits (56), Expect(2) = 0.004
Identities = 13/51 (25%), Positives = 22/51 (42%)
Frame = +2
Query: 106 EYLLRVCVLEEGGAWDSHLPLIEFTYNNSYHSSIGMAPFKALYGRRCRTPL 156
E+L V W S L + + + IG+ PF+ +Y + C P+
Sbjct: 212 EFLREKNVASSRKDWSSKLEDALWACKTAKKTPIGLTPFQMVYRKACHLPV 364
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,073,627
Number of Sequences: 63676
Number of extensions: 204697
Number of successful extensions: 1000
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 980
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 984
length of query: 329
length of database: 12,639,632
effective HSP length: 98
effective length of query: 231
effective length of database: 6,399,384
effective search space: 1478257704
effective search space used: 1478257704
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC130807.4


