

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130807.11 - phase: 0
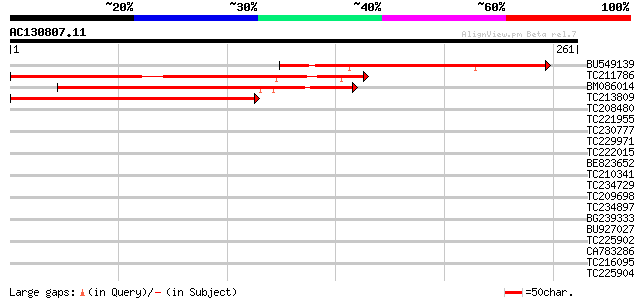
(261 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU549139 162 1e-40
TC211786 138 2e-33
BM086014 135 2e-32
TC213809 129 1e-30
TC208480 37 0.012
TC221955 weakly similar to UP|Q6UAL1 (Q6UAL1) Myosin heavy chain... 33 0.13
TC230777 similar to GB|BAB08884.1|9758397|AB022212 signal recogn... 32 0.22
TC229971 weakly similar to GB|CAB10335.1|2244913|ATFCA4 SEN1 lik... 32 0.22
TC222015 weakly similar to UP|Q9VPS3 (Q9VPS3) CG2839-PA, partial... 32 0.38
BE823652 weakly similar to GP|19699283|gb| At3g51780/ORF3 {Arabi... 30 0.85
TC210341 similar to UP|O64802 (O64802) T1F15.10 protein, partial... 29 1.9
TC234729 29 1.9
TC209698 weakly similar to UP|Q9LS52 (Q9LS52) Gb|AAD39678.1, par... 29 1.9
TC234897 weakly similar to UP|TIR3_YEAST (P40552) Cell wall prot... 28 3.2
BG239333 28 3.2
BU927027 weakly similar to GP|9294370|dbj transcription factor X... 27 7.2
TC225902 similar to UP|O48937 (O48937) NADPH cytochrome P450 red... 27 7.2
CA783286 27 7.2
TC216095 similar to UP|Q761Z4 (Q761Z4) BRI1-KD interacting prote... 27 9.4
TC225904 similar to UP|O48937 (O48937) NADPH cytochrome P450 red... 27 9.4
>BU549139
Length = 534
Score = 162 bits (410), Expect = 1e-40
Identities = 89/160 (55%), Positives = 102/160 (63%), Gaps = 35/160 (21%)
Frame = -2
Query: 125 NSSIRMTRKRMRQEQDALDIEARCIPSENEG----------------------------- 155
+SSIRMTRK +Q AL+ EAR I ENEG
Sbjct: 533 SSSIRMTRKHTKQI--ALEKEARSISLENEGNSVERESTPNACKETVSDNLLECCTKAND 360
Query: 156 ---VDTQESDHQNWLVHALFEYTLDMKLSTDCQTGRLCLSAMHQSSGYSFSISWISRAPG 212
+D ES H NWL+ LFEY L MKLS D Q GR+CLSA+H SSGYSFS+SWI ++PG
Sbjct: 359 RSGIDLHESGHGNWLIQTLFEYALSMKLSLDYQAGRICLSALHGSSGYSFSLSWIGKSPG 180
Query: 213 E---EAELLYHVLSLGTLERLAPEWMREDIMFSPTMCPIF 249
E EAELLYHV+SLGT ER+APEWMREDIMFSP+MCPIF
Sbjct: 179 EEEAEAELLYHVISLGTFERVAPEWMREDIMFSPSMCPIF 60
>TC211786
Length = 591
Score = 138 bits (348), Expect = 2e-33
Identities = 81/173 (46%), Positives = 113/173 (64%), Gaps = 8/173 (4%)
Frame = +3
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
ME LY KLY KYT LK+ KLS+L+ ++KEQE KF+ +++AAE++I+HL++E + L+ Q+N
Sbjct: 54 MEILYSKLYDKYTKLKSKKLSDLDHINKEQEGKFVNYLTAAEELIEHLKSEKEGLVEQVN 233
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVN 120
+ AK+ E+ ++++LL+EEK+KNEAL EEVEKL KL +E TS L N +
Sbjct: 234 E---------AAKEKEVADYQKLLMEEKEKNEALSEEVEKLLKLHQERTSHVLYNNSKIK 386
Query: 121 N-------TSNNSSIRMTRKRMRQEQDALDIEARCIPS-ENEGVDTQESDHQN 165
SN+SSIRMTRKR RQ + +E IPS ENEG + QN
Sbjct: 387 TVDDQFKANSNSSSIRMTRKRTRQ----IALEKETIPSFENEGNSVERESTQN 533
>BM086014
Length = 428
Score = 135 bits (339), Expect = 2e-32
Identities = 76/144 (52%), Positives = 108/144 (74%), Gaps = 6/144 (4%)
Frame = +2
Query: 23 LEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKR 82
L+ +++EQELKF F SAAE++I+HL TE ++LLGQ++ L +EL S+R AKDN++V+++R
Sbjct: 2 LDHLNEEQELKFRNFSSAAEELIEHLMTEKEELLGQVHHLRSELASLRAAKDNQVVDYQR 181
Query: 83 LLLEEKKKNEALFEEVEKLQKLLKEGTSGDLS-NRKVVN-----NTSNNSSIRMTRKRMR 136
LL+EE +KNEAL EEVEKL KL +EGTS DL+ N K++ + +SS+RMTRKR R
Sbjct: 182 LLMEETRKNEALSEEVEKLLKLRQEGTSHDLNYNSKIMTLDDQFKANYSSSVRMTRKRTR 361
Query: 137 QEQDALDIEARCIPSENEGVDTQE 160
Q+AL+ EAR I EN+ ++ E
Sbjct: 362 --QNALEKEARLISFENDQANSVE 427
>TC213809
Length = 458
Score = 129 bits (325), Expect = 1e-30
Identities = 62/115 (53%), Positives = 90/115 (77%)
Frame = +2
Query: 1 MEALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIN 60
ME LY KLY KYT LK+ KLS+L+ ++KEQE+KF+ +++AAE++I+HL++E + LL Q+N
Sbjct: 113 MEILYSKLYDKYTKLKSKKLSDLDHINKEQEVKFVNYLTAAEELIEHLKSEKEGLLEQVN 292
Query: 61 DLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSN 115
+L EL S+R AK+ E+ ++++L +EEK+KNE L EEVEKL KL +E T DL N
Sbjct: 293 ELRTELASLRAAKEKEVADYQKLFMEEKEKNETLSEEVEKLLKLHQERTCHDLYN 457
>TC208480
Length = 1576
Score = 36.6 bits (83), Expect = 0.012
Identities = 29/98 (29%), Positives = 50/98 (50%), Gaps = 9/98 (9%)
Frame = +3
Query: 19 KLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNELV 78
KL E +VH +++ +++ I +L+ ENDKLL ++ L + +++ DNEL
Sbjct: 171 KLKEENEVHIQEDF-------LSKETIKNLKEENDKLLQKVVSLEEVINNLQT--DNELQ 323
Query: 79 EHKRLLLEEK-----KKNEALFEE----VEKLQKLLKE 107
K LE + +N +L ++ VEK +LL E
Sbjct: 324 TQKHTSLEMRIAQLQSENNSLLQKEAGLVEKTNQLLNE 437
>TC221955 weakly similar to UP|Q6UAL1 (Q6UAL1) Myosin heavy chain class XI E3
protein, partial (6%)
Length = 728
Score = 33.1 bits (74), Expect = 0.13
Identities = 36/137 (26%), Positives = 58/137 (42%), Gaps = 12/137 (8%)
Frame = +1
Query: 15 LKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVR---- 70
L TNK ELE +E + K +F E+ + EN L + + + T ++
Sbjct: 166 LLTNKNEELETEVEELKKKIKEF----EESYSEIENENQARLKEAEEAQLKATQLQETIE 333
Query: 71 --------VAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKVVNNT 122
+ +N+++ K L E+ KNE LFEE+ K+LK+ ++ N
Sbjct: 334 RLELSLSNLESENQVLCQKAL---EEPKNEELFEEI----KILKD---------QIANLQ 465
Query: 123 SNNSSIRMTRKRMRQEQ 139
S N S+R EQ
Sbjct: 466 SENESLRSQAAAAALEQ 516
>TC230777 similar to GB|BAB08884.1|9758397|AB022212 signal recognition
particle 68kD protein-like {Arabidopsis thaliana;} ,
partial (47%)
Length = 1077
Score = 32.3 bits (72), Expect = 0.22
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 8/65 (12%)
Frame = +1
Query: 51 ENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEV--------EKLQ 102
+N ++ ++ DL NE S N +EH ++EEKK E L E V E+L+
Sbjct: 574 DNKTMMKELEDLCNECRS------NSCIEHALGIMEEKKTQENLSERVSNISLTGTERLE 735
Query: 103 KLLKE 107
K L E
Sbjct: 736 KFLLE 750
>TC229971 weakly similar to GB|CAB10335.1|2244913|ATFCA4 SEN1 like protein
{Arabidopsis thaliana;} , partial (14%)
Length = 919
Score = 32.3 bits (72), Expect = 0.22
Identities = 33/126 (26%), Positives = 47/126 (37%), Gaps = 23/126 (18%)
Frame = +1
Query: 4 LYKKLYSKYTTLKTNKLSELEDVHKEQELKFLK-----FVSAAEDVID------------ 46
LY+KL S Y TLK+ + + +Q F K F +AE V+D
Sbjct: 55 LYQKLISLYPTLKSGNQVAIISPYSQQVKLFQKRFEETFGMSAEKVVDICTVDGCQGREK 234
Query: 47 ------HLRTENDKLLGQINDLGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEK 100
+R DK +G + D+ + AK LV L ++ L E EK
Sbjct: 235 DIAIFSCVRASKDKGIGFVEDIRRMNVGITRAKSAVLVVGSASTLRRSEQWNKLVESAEK 414
Query: 101 LQKLLK 106
L K
Sbjct: 415 RNCLFK 432
>TC222015 weakly similar to UP|Q9VPS3 (Q9VPS3) CG2839-PA, partial (4%)
Length = 796
Score = 31.6 bits (70), Expect = 0.38
Identities = 33/154 (21%), Positives = 61/154 (39%), Gaps = 10/154 (6%)
Frame = +3
Query: 2 EALYKKLYSKYTTLKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQIND 61
E+L K+++ +Y + E E KE+ K K E R E + + D
Sbjct: 141 ESLCKEVFEEYIAQLKEEAKESERKRKEERAKKEKDREERERRKGKQRKEKEGGRERGKD 320
Query: 62 LGN----------ELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSG 111
+ ELT ++ +K+N+ E +K+++ + EK +K G+
Sbjct: 321 EAHKKDKADSDSMELTEIQTSKENKRSEDDNRKQRKKRQSPVYEMDKEKTKKSHGHGSDS 500
Query: 112 DLSNRKVVNNTSNNSSIRMTRKRMRQEQDALDIE 145
S R + S+ + ++ R+E D D E
Sbjct: 501 KKSRRHASGHESDEGRHKRHKRDHRREGDLEDGE 602
>BE823652 weakly similar to GP|19699283|gb| At3g51780/ORF3 {Arabidopsis
thaliana}, partial (31%)
Length = 742
Score = 30.4 bits (67), Expect = 0.85
Identities = 22/82 (26%), Positives = 40/82 (47%)
Frame = -3
Query: 18 NKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKDNEL 77
+K +LE++HK ++ +S A + I +RTE DKL ++ L + +D E
Sbjct: 590 SKERKLEEMHKSED------ISKACEAISKVRTEVDKLYQKVVALETTVCGGAKVEDKEF 429
Query: 78 VEHKRLLLEEKKKNEALFEEVE 99
LL+ + K +++ E E
Sbjct: 428 AILTELLMVQLLKLDSIAAEGE 363
>TC210341 similar to UP|O64802 (O64802) T1F15.10 protein, partial (16%)
Length = 777
Score = 29.3 bits (64), Expect = 1.9
Identities = 14/44 (31%), Positives = 23/44 (51%)
Frame = -2
Query: 126 SSIRMTRKRMRQEQDALDIEARCIPSENEGVDTQESDHQNWLVH 169
++IR RKR R+ + A + ARC EN+ + E+ + H
Sbjct: 404 AAIRRQRKRRRRRRGAAEAAARCRSIENDDANDMENRKMEQMNH 273
>TC234729
Length = 476
Score = 29.3 bits (64), Expect = 1.9
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +3
Query: 162 DHQNWLVHALFEYTLDMKLS 181
DH NWL++ + Y LD +LS
Sbjct: 66 DHSNWLLYTIVSYDLDSRLS 125
>TC209698 weakly similar to UP|Q9LS52 (Q9LS52) Gb|AAD39678.1, partial (41%)
Length = 1050
Score = 29.3 bits (64), Expect = 1.9
Identities = 25/89 (28%), Positives = 43/89 (48%), Gaps = 7/89 (7%)
Frame = +2
Query: 88 KKKNEALFE-----EVEKLQKLLKEGTSGDLSNR--KVVNNTSNNSSIRMTRKRMRQEQD 140
+ +N A+F E+EK++KLL+E T G L + + + N SS R + ++ ++
Sbjct: 86 RPRNRAVFSGFTNAEIEKMEKLLREPTGGSLGREFYQKLARSFNYSSGRAGKPIIKWTEE 265
Query: 141 ALDIEARCIPSENEGVDTQESDHQNWLVH 169
L + S N D Q+ +H L H
Sbjct: 266 KL---CKTHQSWNLKQDHQKMEHGMMLRH 343
>TC234897 weakly similar to UP|TIR3_YEAST (P40552) Cell wall protein TIR3
precursor, partial (12%)
Length = 698
Score = 28.5 bits (62), Expect = 3.2
Identities = 24/58 (41%), Positives = 32/58 (54%), Gaps = 3/58 (5%)
Frame = +3
Query: 56 LGQIND---LGNELTSVRVAKDNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTS 110
+GQ+ND NEL S A+DN +K++++EE V L KLLKEGTS
Sbjct: 408 MGQLNDRIEAANELASF--AQDN--ARNKKIIVEECG--------VPPLLKLLKEGTS 545
>BG239333
Length = 376
Score = 28.5 bits (62), Expect = 3.2
Identities = 22/78 (28%), Positives = 41/78 (52%)
Frame = +2
Query: 15 LKTNKLSELEDVHKEQELKFLKFVSAAEDVIDHLRTENDKLLGQINDLGNELTSVRVAKD 74
L+ +LS + + ++ + + +S A V+ LR E +L ++L ++ ++ K
Sbjct: 38 LRFLELSSILEPSRQPKSDKVAILSDAARVVIQLRNEAKRLKEMNDELQAKVKELKGEK- 214
Query: 75 NELVEHKRLLLEEKKKNE 92
NEL + K L EEK+K E
Sbjct: 215 NELRDEKNRLKEEKEKLE 268
>BU927027 weakly similar to GP|9294370|dbj transcription factor X1-like
protein {Arabidopsis thaliana}, partial (18%)
Length = 443
Score = 27.3 bits (59), Expect = 7.2
Identities = 21/95 (22%), Positives = 45/95 (47%), Gaps = 5/95 (5%)
Frame = +1
Query: 51 ENDKLLGQINDLGNELTSVRVAKDNEL----VEHKRLLLEEK-KKNEALFEEVEKLQKLL 105
E DKL+ N E+ ++ + + L +H++L L+ K +KNE +VE ++
Sbjct: 13 EKDKLIQAYNA---EIKKIQSSATDHLKKIFTDHEKLKLQLKSQKNELELRKVELEKREA 183
Query: 106 KEGTSGDLSNRKVVNNTSNNSSIRMTRKRMRQEQD 140
+ ++++ N NSS++M ++ +
Sbjct: 184 HNESERKKLTKEIMENAMKNSSLQMATLEQKKADE 288
>TC225902 similar to UP|O48937 (O48937) NADPH cytochrome P450 reductase ,
partial (11%)
Length = 581
Score = 27.3 bits (59), Expect = 7.2
Identities = 12/35 (34%), Positives = 19/35 (54%)
Frame = -2
Query: 195 HQSSGYSFSISWISRAPGEEAELLYHVLSLGTLER 229
H + S +I++ ++YH LSLGT+ER
Sbjct: 250 HPEACIQISSPYIAQISASSL*IVYHALSLGTIER 146
>CA783286
Length = 460
Score = 27.3 bits (59), Expect = 7.2
Identities = 13/31 (41%), Positives = 20/31 (63%), Gaps = 5/31 (16%)
Frame = +2
Query: 88 KKKNEALFE-----EVEKLQKLLKEGTSGDL 113
+ +N A+F E+EK++KLL+E T G L
Sbjct: 98 RPRNRAVFSGFTNAEIEKMEKLLREPTGGSL 190
>TC216095 similar to UP|Q761Z4 (Q761Z4) BRI1-KD interacting protein 120
(Fragment), partial (13%)
Length = 914
Score = 26.9 bits (58), Expect = 9.4
Identities = 18/45 (40%), Positives = 27/45 (60%)
Frame = +1
Query: 74 DNELVEHKRLLLEEKKKNEALFEEVEKLQKLLKEGTSGDLSNRKV 118
D E +E KRL E+K + + E+ KLQ + +S DLS++KV
Sbjct: 304 DVEYLERKRLKKEKKHREK----ELAKLQSDEAKRSSIDLSSKKV 426
>TC225904 similar to UP|O48937 (O48937) NADPH cytochrome P450 reductase ,
partial (68%)
Length = 1704
Score = 26.9 bits (58), Expect = 9.4
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = -1
Query: 203 SISWISRAPGEEAELLYHVLSLGTLER 229
S +I++ +LYH LSLGT+ER
Sbjct: 1428 SSPYIAQISASSL*ILYHALSLGTIER 1348
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.131 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,121,726
Number of Sequences: 63676
Number of extensions: 129906
Number of successful extensions: 509
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 500
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 502
length of query: 261
length of database: 12,639,632
effective HSP length: 95
effective length of query: 166
effective length of database: 6,590,412
effective search space: 1094008392
effective search space used: 1094008392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC130807.11


