

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130806.14 - phase: 0
(394 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
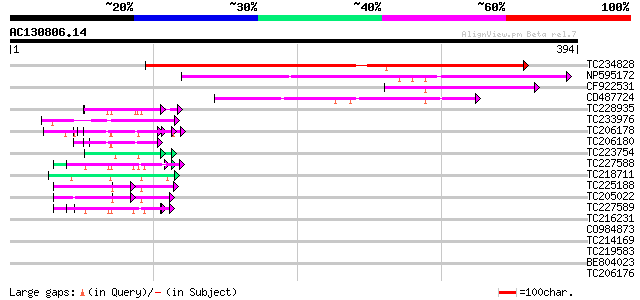
Score E
Sequences producing significant alignments: (bits) Value
TC234828 225 2e-59
NP595172 polyprotein [Glycine max] 81 7e-16
CF922531 60 1e-09
CD487724 58 8e-09
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 54 2e-07
TC233976 53 2e-07
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 51 8e-07
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 49 3e-06
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 49 5e-06
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 49 5e-06
TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), ... 48 9e-06
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 47 1e-05
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 47 1e-05
TC227589 47 1e-05
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 39 0.005
CO984873 35 0.045
TC214169 similar to UP|Q8LF25 (Q8LF25) DNA-damage inducible prot... 35 0.059
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 33 0.17
BE804023 33 0.29
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 28 7.2
>TC234828
Length = 857
Score = 225 bits (574), Expect = 2e-59
Identities = 116/270 (42%), Positives = 167/270 (60%), Gaps = 4/270 (1%)
Frame = +3
Query: 95 NCNGEGHISSQCTQPKKVRTGGKVFALAGMQTTNEDRLIRGTCFFNSIPLIAIIDTGATH 154
N G++S+ + + +VFA++G + D LIRG C L + D+GATH
Sbjct: 66 NSTRSGNVSNNNISGGRPKVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATH 245
Query: 155 CFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLICPISMFGRDFEVDLVCLPLT 214
FI+ C +LGL +++ +MVV TP VTTS VCL CPI + GR F DL+CLPL
Sbjct: 246 SFISHACVERLGLCATELPYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLA 425
Query: 215 GMDVIFGMNWLEYNRVHINCFNKTVHFSSAEEESGVEILSTKEMKQ----LERDGILMYS 270
+DVI GM+WL N + ++C K + F G +++ ++ +K+ E + + Y
Sbjct: 426 HLDVILGMDWLSTNHIFLDCKEKMLVF-------GGDVVPSEPLKEDAANEETEDVRTYM 584
Query: 271 LMACLSLENQAVIDKLPVVNEFPEVFSDEIPDVPPEREVEFSIDLVPRTKPVSMAPYRMS 330
++ + +E A + +PVV+EFPEVF D++ ++PPEREVEF ID+VP PVS+APYRMS
Sbjct: 585 VLFSMYVEEDAEVSCIPVVSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMS 764
Query: 331 ASELAELKNQLEDLLDKKFVRASVFPWGAP 360
ELAE+K Q++DLL K+FVR S PWGAP
Sbjct: 765 PVELAEVKAQVQDLLSKQFVRPSASPWGAP 854
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 81.3 bits (199), Expect = 7e-16
Identities = 70/292 (23%), Positives = 135/292 (45%), Gaps = 21/292 (7%)
Frame = +1
Query: 120 ALAGMQTTNEDRLIRGTCFFNSIPLIAIIDTGATHCFIALECAYKLGLIVSDMKGEMVVE 179
+L M+ +N IR T I + ++D G++ FI A L L V V+
Sbjct: 1132 SLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAPNLRVLV 1311
Query: 180 TPAKGSVTTSLVCLICPISMFGRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNK-T 238
+ +V + P+ + G++ +V + L ++G DVI G WL H+ + T
Sbjct: 1312 GNGQILSAEGIVQQL-PLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADYAALT 1488
Query: 239 VHFSSAEEESGVEILSTKEMKQLERDGILMY----SLMACLSLE------NQAVIDKLP- 287
+ F ++ ++ E Q + S+ C +++ + + LP
Sbjct: 1489 LKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKEVPEDTLKDLPT 1668
Query: 288 --------VVNEFPEVFSDEIP-DVPPEREVEFSIDLVPRTKPVSMAPYRMSASELAELK 338
+++ + +VF+ +P +PP+RE + +I L + PV + PYR ++ +++
Sbjct: 1669 NIDPELAILLHTYAQVFA--VPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIE 1842
Query: 339 NQLEDLLDKKFVRASVFPWGAPVLLAKKKDGSMRLCIAYPPLNKVPIKNRYP 390
++++L + ++ S P+ P+LL KKKDGS R C Y LN + +K+ +P
Sbjct: 1843 KMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFP 1998
>CF922531
Length = 602
Score = 60.5 bits (145), Expect = 1e-09
Identities = 40/114 (35%), Positives = 63/114 (55%), Gaps = 6/114 (5%)
Frame = -2
Query: 261 LERDGILMYSLMACLSLENQAVIDKLP-----VVNEFPEVFSDEIP-DVPPEREVEFSID 314
L++ L+ S LS I+ LP +++EF ++F EIP +PP R +E ID
Sbjct: 343 LKQSFYLLLSRETSLSTVTIPTIETLPPKVQELLHEFGDIFPKEIPLGLPPLRGIEHQID 164
Query: 315 LVPRTKPVSMAPYRMSASELAELKNQLEDLLDKKFVRASVFPWGAPVLLAKKKD 368
LVPR + YR + E E+++Q+++LL+K +V+ S+ VLL KKD
Sbjct: 163 LVPRASLPNRPTYRTNPQETKEIESQVKELLEKGWVQESLSLCVVLVLLVPKKD 2
>CD487724
Length = 676
Score = 57.8 bits (138), Expect = 8e-09
Identities = 49/202 (24%), Positives = 93/202 (45%), Gaps = 17/202 (8%)
Frame = +3
Query: 143 PLIAIIDTGATHCFIALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLICPISMFGR 202
PL+ ++D G+TH F+ +LGL V+ T+ +C PIS+
Sbjct: 21 PLVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTT-ICEAIPISIQNI 197
Query: 203 DFEVDLVCLPLTGMDVIFGMNWL--------EYNRVHINCF--NKTVHFSSAEEESGVEI 252
+F V L LP+ G +++ G+ WL +YN + + F ++ V E E+ + +
Sbjct: 198 EFLVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQL-KGESEAQLGL 374
Query: 253 LSTKEMKQLERDGILMYSLMACLSLENQAVIDKLP-------VVNEFPEVFSDEIPDVPP 305
L+ ++++L + + + EN + P ++++F +F +PP
Sbjct: 375 LNHHQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFSALFQ*P-QGLPP 551
Query: 306 EREVEFSIDLVPRTKPVSMAPY 327
RE + I L+P ++PV+M Y
Sbjct: 552 ARETDHHIHLLP*SEPVNMRLY 617
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 53.5 bits (127), Expect = 2e-07
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 13/70 (18%)
Frame = +1
Query: 52 VCFKCGEKGHKSNVC------TKDEKKCFRCGQKGHVLADC----KRGD---IVCYNCNG 98
+C +C +GH++ C KD K C+ CG+ GH L C + G C+ CN
Sbjct: 343 ICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQEGGTKFAECFVCNQ 522
Query: 99 EGHISSQCTQ 108
GH+S C Q
Sbjct: 523 RGHLSKNCPQ 552
Score = 45.1 bits (105), Expect = 6e-05
Identities = 25/79 (31%), Positives = 35/79 (43%), Gaps = 11/79 (13%)
Frame = +1
Query: 53 CFKCGEKGHKSNVCTKD-----EKKCFRCGQKGHVLADCK------RGDIVCYNCNGEGH 101
CF C H + +C + K C RC ++GH +C + CYNC GH
Sbjct: 271 CFICKAMDHIAKLCPEKAEWEKNKICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGH 450
Query: 102 ISSQCTQPKKVRTGGKVFA 120
+QC P ++ GG FA
Sbjct: 451 ALTQCLHP--LQEGGTKFA 501
Score = 28.1 bits (61), Expect = 7.2
Identities = 13/41 (31%), Positives = 18/41 (43%), Gaps = 7/41 (17%)
Frame = +1
Query: 53 CFKCGEKGHKSNVCTKDEKK-------CFRCGQKGHVLADC 86
CF C ++GH S C ++ C CG H+ DC
Sbjct: 505 CFVCNQRGHLSKNCPQNTHGIYPKGGCCKICGGVTHLAKDC 627
>TC233976
Length = 763
Score = 53.1 bits (126), Expect = 2e-07
Identities = 33/114 (28%), Positives = 49/114 (42%), Gaps = 18/114 (15%)
Frame = +1
Query: 23 KPYSAP------------------AGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSN 64
KPYSAP AG Q +N +P GR G G +
Sbjct: 106 KPYSAPLEYENHYGGQRTSGGHHLAGGSSQLVNRVSQPAGR-----------GGSGAPAI 252
Query: 65 VCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCTQPKKVRTGGKV 118
V T +C +CG+ GH +C ++ C+N G+GH+S+ C P+K + G +
Sbjct: 253 VTTP--LRCRKCGRLGHNAHECTYREVTCFNYQGKGHLSTNCPHPRKEKRSGSL 408
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 51.2 bits (121), Expect = 8e-07
Identities = 27/91 (29%), Positives = 46/91 (49%), Gaps = 5/91 (5%)
Frame = +2
Query: 24 PYSAPAGKGKQRLN---DERRPK--GRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQ 78
PY + +G R N + +RP R+ P +C CG GH ++ CT + C+ C +
Sbjct: 299 PYRRDSRRGFSRDNLCKNCKRPGHYARECPNVAICHNCGLPGHIASECT-TKSLCWNCKE 475
Query: 79 KGHVLADCKRGDIVCYNCNGEGHISSQCTQP 109
GH+ + C + +C+ C GH + +C+ P
Sbjct: 476 PGHMASSCP-NEGICHTCGKAGHRARECSAP 565
Score = 51.2 bits (121), Expect = 8e-07
Identities = 27/77 (35%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Frame = +2
Query: 52 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKR-----GDI-VCYNCNGEGHISSQ 105
+C+ C E GH ++ C +E C CG+ GH +C GD+ +C NC +GHI+++
Sbjct: 455 LCWNCKEPGHMASSCP-NEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAE 631
Query: 106 CTQPKKVRTGGKVFALA 122
CT K K LA
Sbjct: 632 CTNEKACNNCRKTGHLA 682
Score = 50.4 bits (119), Expect = 1e-06
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 2/67 (2%)
Frame = +2
Query: 52 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCTQPKK 111
+C C ++GH + CT +EK C C + GH+ DC D +C CN GH++ QC +
Sbjct: 590 LCNNCYKQGHIAAECT-NEKACNNCRKTGHLARDCP-NDPICNLCNVSGHVARQCPKANV 763
Query: 112 V--RTGG 116
+ R+GG
Sbjct: 764 LGDRSGG 784
Score = 49.3 bits (116), Expect = 3e-06
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 6/65 (9%)
Frame = +2
Query: 48 PAEIVCFKCGEKGHKSNVCTK------DEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGH 101
P E +C CG+ GH++ C+ D + C C ++GH+ A+C + C NC GH
Sbjct: 500 PNEGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC-TNEKACNNCRKTGH 676
Query: 102 ISSQC 106
++ C
Sbjct: 677 LARDC 691
Score = 46.2 bits (108), Expect = 3e-05
Identities = 23/87 (26%), Positives = 34/87 (38%), Gaps = 25/87 (28%)
Frame = +2
Query: 45 RDTPAEIVCFKCGEKGHKSNVCTKD-------------------------EKKCFRCGQK 79
RD P + +C C GH + C K + C C Q
Sbjct: 683 RDCPNDPICNLCNVSGHVARQCPKANVLGDRSGGGGGGGGARGGGGGGYRDVVCRNCQQL 862
Query: 80 GHVLADCKRGDIVCYNCNGEGHISSQC 106
GH+ DC ++C+NC G GH++ +C
Sbjct: 863 GHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 37.0 bits (84), Expect = 0.015
Identities = 14/40 (35%), Positives = 20/40 (50%)
Frame = +2
Query: 50 EIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRG 89
++VC C + GH S C C CG +GH+ +C G
Sbjct: 833 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 952
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 49.3 bits (116), Expect = 3e-06
Identities = 22/55 (40%), Positives = 32/55 (58%)
Frame = +3
Query: 52 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQC 106
+C C ++GH + CT +EK C C + GH+ DC D +C CN GH++ QC
Sbjct: 75 LCNNCYKQGHIAAECT-NEKACNNCRKTGHLARDCP-NDPICNLCNVSGHVARQC 233
Score = 47.4 bits (111), Expect = 1e-05
Identities = 23/84 (27%), Positives = 34/84 (40%), Gaps = 22/84 (26%)
Frame = +3
Query: 45 RDTPAEIVCFKCGEKGHKSNVCTKD----------------------EKKCFRCGQKGHV 82
RD P + +C C GH + C K + C C Q GH+
Sbjct: 168 RDCPNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGGYRDVVCRNCQQLGHM 347
Query: 83 LADCKRGDIVCYNCNGEGHISSQC 106
DC ++C+NC G GH++ +C
Sbjct: 348 SRDCMGPLMICHNCGGRGHLAYEC 419
Score = 43.1 bits (100), Expect = 2e-04
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 6/57 (10%)
Frame = +3
Query: 56 CGEKGHKSNVCTK------DEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQC 106
CG+ GH++ C+ D + C C ++GH+ A+C + C NC GH++ C
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAEC-TNEKACNNCRKTGHLARDC 176
Score = 37.0 bits (84), Expect = 0.015
Identities = 14/40 (35%), Positives = 20/40 (50%)
Frame = +3
Query: 50 EIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRG 89
++VC C + GH S C C CG +GH+ +C G
Sbjct: 309 DVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 428
Score = 36.2 bits (82), Expect = 0.026
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Frame = +3
Query: 76 CGQKGHVLADCKR-----GDI-VCYNCNGEGHISSQCTQPKKVRTGGKVFALA 122
CG+ GH +C GD+ +C NC +GHI+++CT K K LA
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEKACNNCRKTGHLA 167
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 48.5 bits (114), Expect = 5e-06
Identities = 26/86 (30%), Positives = 34/86 (39%), Gaps = 22/86 (25%)
Frame = +2
Query: 53 CFKCGEKGHKSNVCTKDEKK----------CFRCGQKGHVLADCKR------------GD 90
C++CGE GH + C + CF CG GH+ DC R G
Sbjct: 44 CYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCVRGGGGSVGIGGGGGG 223
Query: 91 IVCYNCNGEGHISSQCTQPKKVRTGG 116
C+ C G GH++ C K GG
Sbjct: 224 GSCFRCGGFGHMARDCATGKGNIGGG 301
Score = 47.8 bits (112), Expect = 9e-06
Identities = 23/80 (28%), Positives = 32/80 (39%), Gaps = 24/80 (30%)
Frame = +2
Query: 53 CFKCGEKGHKSNVCTKDEKK---------CFRCGQKGHVLADCKR--------------- 88
CF+CG GH + C + CFRCG+ GH+ DC
Sbjct: 230 CFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDCGMEGGRFGGGGGSGGGG 409
Query: 89 GDIVCYNCNGEGHISSQCTQ 108
G C+NC GH + +C +
Sbjct: 410 GKSTCFNCGKPGHFARECVE 469
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 48.5 bits (114), Expect = 5e-06
Identities = 29/85 (34%), Positives = 37/85 (43%), Gaps = 13/85 (15%)
Frame = +1
Query: 40 RRPKGRDTPAEIVCFKCGEKGHKSNVCTKD----EKKCFRCGQKGHVLADCKR--GDIV- 92
R +D EI C+ C GH V T D E C++CGQ GH C R +I
Sbjct: 307 RNDYSQDDLKEIQCYVCKRLGHLCCVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITS 486
Query: 93 ------CYNCNGEGHISSQCTQPKK 111
C+ C EGH + +CT K
Sbjct: 487 GATPSSCFKCGEEGHFARECTSSIK 561
Score = 48.1 bits (113), Expect = 7e-06
Identities = 28/97 (28%), Positives = 39/97 (39%), Gaps = 11/97 (11%)
Frame = +1
Query: 31 KGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKD-------EKKCFRCGQKGHVL 83
KG R D + + +C KCG GH C D E +C+ C + GH+
Sbjct: 199 KGGHRAKDCPEKHTSTSKSIAICLKCGNSGHDIFSCRNDYSQDDLKEIQCYVCKRLGHLC 378
Query: 84 A----DCKRGDIVCYNCNGEGHISSQCTQPKKVRTGG 116
D G+I CY C GH C++ + T G
Sbjct: 379 CVNTDDATPGEISCYKCGQLGHTGLACSRLRDEITSG 489
Score = 47.0 bits (110), Expect = 1e-05
Identities = 29/86 (33%), Positives = 41/86 (46%), Gaps = 4/86 (4%)
Frame = +1
Query: 40 RRPKGRDTPAEI--VCFKCGEKGHKSNVCT--KDEKKCFRCGQKGHVLADCKRGDIVCYN 95
R P+ D P CF CGE+GH + C+ K +K C+ CG GH C + C+
Sbjct: 16 RGPRYFDPPDNSWGACFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQCSKVQ-DCFI 192
Query: 96 CNGEGHISSQCTQPKKVRTGGKVFAL 121
C GH + C P+K + K A+
Sbjct: 193 CKKGGHRAKDC--PEKHTSTSKSIAI 264
Score = 32.3 bits (72), Expect = 0.38
Identities = 20/52 (38%), Positives = 24/52 (45%)
Frame = +1
Query: 30 GKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGH 81
G RL DE TP+ CFKCGE+GH + CT K R + H
Sbjct: 448 GLACSRLRDEITSGA--TPSS--CFKCGEEGHFARECTSSIKSGKRNWESSH 591
>TC218711 similar to UP|Q90Z60 (Q90Z60) Rev-Erb beta (Fragment), partial (6%)
Length = 1204
Score = 47.8 bits (112), Expect = 9e-06
Identities = 33/113 (29%), Positives = 44/113 (38%), Gaps = 22/113 (19%)
Frame = -3
Query: 28 PAGKGKQRLNDERR----PKGRDTPAEIVCFKCGEKGHKSNVCTK------DEKKCFRCG 77
PAGK ++ D R P + + C C + GH+ C + DE+ CF CG
Sbjct: 608 PAGKTLVQITDTLRVTKSPFKHHGESSLRCRACRQPGHRFQQCQRLKCLSMDEEVCFFCG 429
Query: 78 QKGHVLADCKRGD------IVCYNCNGEGHISSQCTQ------PKKVRTGGKV 118
+ GH L C C C G GH S C Q PK + G +
Sbjct: 428 EIGHSLGKCDVSQAGGGRFAKCLLCYGHGHFSYNCPQNGHGIDPKVLAVNGAI 270
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 47.0 bits (110), Expect = 1e-05
Identities = 22/49 (44%), Positives = 25/49 (50%), Gaps = 3/49 (6%)
Frame = +1
Query: 72 KCFRCGQKGHVLADCKRGD--IVCYNCNGEGHISSQC-TQPKKVRTGGK 117
+CF CG GH DCK GD CY C GHI C PKK+ G+
Sbjct: 361 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKLSQRGR 507
Score = 45.8 bits (107), Expect = 3e-05
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Frame = +1
Query: 31 KGKQRLNDERRPKGRDTP-AEIVCFKCGEKGHKSNVCTKDE--KKCFRCGQKGHVLADCK 87
KG R + +R GR P CF CG GH + C + KC+RCG++GH+ +CK
Sbjct: 295 KGGPRGSRDREYMGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCK 474
Score = 29.6 bits (65), Expect = 2.5
Identities = 10/27 (37%), Positives = 16/27 (59%)
Frame = +1
Query: 53 CFKCGEKGHKSNVCTKDEKKCFRCGQK 79
C++CGE+GH C KK + G++
Sbjct: 430 CYRCGERGHIEKNCKNSPKKLSQRGRR 510
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 47.0 bits (110), Expect = 1e-05
Identities = 22/46 (47%), Positives = 24/46 (51%), Gaps = 3/46 (6%)
Frame = +2
Query: 72 KCFRCGQKGHVLADCKRGD--IVCYNCNGEGHISSQC-TQPKKVRT 114
+CF CG GH DCK GD CY C GHI C PKK+ T
Sbjct: 311 RCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCKNSPKKLST 448
Score = 43.5 bits (101), Expect = 2e-04
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Frame = +2
Query: 31 KGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDE--KKCFRCGQKGHVLADCK 87
KG R + E +G P CF CG GH + C + KC+RCG++GH+ +CK
Sbjct: 251 KGAPRGSREYLGRG-PPPGSGRCFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCK 424
Score = 28.1 bits (61), Expect = 7.2
Identities = 9/20 (45%), Positives = 12/20 (60%)
Frame = +2
Query: 53 CFKCGEKGHKSNVCTKDEKK 72
C++CGE+GH C KK
Sbjct: 380 CYRCGERGHIERNCKNSPKK 439
>TC227589
Length = 547
Score = 47.0 bits (110), Expect = 1e-05
Identities = 26/89 (29%), Positives = 36/89 (40%), Gaps = 11/89 (12%)
Frame = +2
Query: 31 KGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKD-------EKKCFRCGQKGHVL 83
KG R D + + +C KCG GH C D E +C+ C + GH+
Sbjct: 185 KGGHRAKDCLEKHTSRSKSVAICLKCGNSGHDMFSCRNDYSPDDLKEIQCYVCKRVGHLC 364
Query: 84 A----DCKRGDIVCYNCNGEGHISSQCTQ 108
D G+I CY C GH C++
Sbjct: 365 CVNTDDATPGEISCYKCGQLGHTGLACSK 451
Score = 46.6 bits (109), Expect = 2e-05
Identities = 26/79 (32%), Positives = 36/79 (44%), Gaps = 4/79 (5%)
Frame = +2
Query: 40 RRPKGRDTPAEI--VCFKCGEKGHKSNVCT--KDEKKCFRCGQKGHVLADCKRGDIVCYN 95
R P+ D P CF CGE GH + C+ K +K C+ CG GH C + C+
Sbjct: 2 RGPRYFDPPDSSWGACFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQCTKAQ-DCFI 178
Query: 96 CNGEGHISSQCTQPKKVRT 114
C GH + C + R+
Sbjct: 179 CKKGGHRAKDCLEKHTSRS 235
Score = 45.4 bits (106), Expect = 4e-05
Identities = 28/77 (36%), Positives = 34/77 (43%), Gaps = 13/77 (16%)
Frame = +2
Query: 46 DTPAEIVCFKCGEKGHKSNVCTKD----EKKCFRCGQKGHV-LADCKRGDIV-------- 92
D EI C+ C GH V T D E C++CGQ GH LA K D +
Sbjct: 311 DDLKEIQCYVCKRVGHLCCVNTDDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSS 490
Query: 93 CYNCNGEGHISSQCTQP 109
C C GH + +CT P
Sbjct: 491 CCKCGEAGHFAQECTSP 541
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 38.5 bits (88), Expect = 0.005
Identities = 15/47 (31%), Positives = 21/47 (43%), Gaps = 13/47 (27%)
Frame = +3
Query: 73 CFRCGQKGHVLADCKR-------------GDIVCYNCNGEGHISSQC 106
C++CG+ GH+ DC + G CYNC GH + C
Sbjct: 231 CYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDC 371
>CO984873
Length = 754
Score = 35.4 bits (80), Expect = 0.045
Identities = 16/41 (39%), Positives = 20/41 (48%), Gaps = 11/41 (26%)
Frame = -2
Query: 68 KDEKKCFRCGQKGHVLADCKR-----------GDIVCYNCN 97
K+E KCF C +KGH+ DC + D VCY N
Sbjct: 372 KNESKCFFCNKKGHIKKDCSKFKSWLNKKNIPFDFVCYESN 250
>TC214169 similar to UP|Q8LF25 (Q8LF25) DNA-damage inducible protein
DDI1-like, partial (90%)
Length = 1604
Score = 35.0 bits (79), Expect = 0.059
Identities = 23/107 (21%), Positives = 47/107 (43%), Gaps = 1/107 (0%)
Frame = +2
Query: 140 NSIPLIAIIDTGATHCFIALECAYKLGLI-VSDMKGEMVVETPAKGSVTTSLVCLICPIS 198
N +PL A +D+GA I+ CA +LGL+ + D + + + + + + PI
Sbjct: 767 NGVPLKAFVDSGAQSTIISKSCAERLGLLRLLDQRYRGIAHGVGQSEILGRI--HVAPIK 940
Query: 199 MFGRDFEVDLVCLPLTGMDVIFGMNWLEYNRVHINCFNKTVHFSSAE 245
+ + + L M+ +FG++ L ++ I+ + E
Sbjct: 941 IGSIFYPCSFLVLDSPNMEFLFGLDMLRKHQCIIDLKENVLRVGGGE 1081
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 33.5 bits (75), Expect = 0.17
Identities = 15/55 (27%), Positives = 22/55 (39%), Gaps = 20/55 (36%)
Frame = +2
Query: 72 KCFRCGQKGHVLADCKRGD--------------------IVCYNCNGEGHISSQC 106
+C+ CG+ GH+ DC G C+NC EGH + +C
Sbjct: 422 ECYNCGRIGHLARDCYHGQGGGGGDDGRNRRRGGGGGGGGGCFNCGEEGHFAREC 586
>BE804023
Length = 407
Score = 32.7 bits (73), Expect = 0.29
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Frame = -3
Query: 53 CFKCGEKGHKSNVCTK-DEKKCFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCTQPKK 111
C C +KGH C K + KC +C Q GH + ++C N N + +++ PK+
Sbjct: 219 C*HCRKKGHPPFRC*KRPDAKCSQCNQMGHEV-------VICKNRNQSHNEAAKVVDPKE 61
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 28.1 bits (61), Expect = 7.2
Identities = 8/17 (47%), Positives = 11/17 (64%)
Frame = +3
Query: 73 CFRCGQKGHVLADCKRG 89
C+ CG+ GH+ DC G
Sbjct: 435 CYNCGESGHLARDCSGG 485
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,877,051
Number of Sequences: 63676
Number of extensions: 290285
Number of successful extensions: 1567
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1383
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1495
length of query: 394
length of database: 12,639,632
effective HSP length: 99
effective length of query: 295
effective length of database: 6,335,708
effective search space: 1869033860
effective search space used: 1869033860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC130806.14


