

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC128638.4 - phase: 0
(561 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
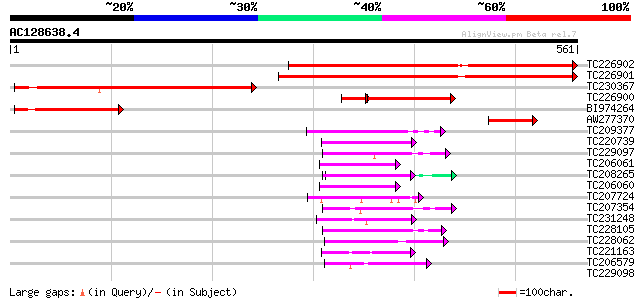
Score E
Sequences producing significant alignments: (bits) Value
TC226902 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia myo... 524 e-149
TC226901 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia myo... 515 e-146
TC230367 379 e-105
TC226900 similar to UP|DMWD_HUMAN (Q09019) Dystrophia myotonica-... 169 3e-42
BI974264 149 4e-36
AW277370 81 1e-15
TC209377 similar to UP|Q9FLX9 (Q9FLX9) Notchless protein homolog... 53 3e-07
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 49 6e-06
TC229097 similar to PIR|T46032|T46032 WD-40 repeat regulatory pr... 48 1e-05
TC206061 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-... 47 2e-05
TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%) 47 2e-05
TC206060 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-... 45 9e-05
TC207724 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] ... 45 1e-04
TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 45 1e-04
TC231248 44 1e-04
TC228105 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {A... 43 3e-04
TC228062 weakly similar to UP|Q39221 (Q39221) St12p protein, par... 43 4e-04
TC221163 42 6e-04
TC206579 weakly similar to UP|TAF5_YEAST (P38129) Transcription ... 42 7e-04
TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory pr... 42 0.001
>TC226902 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia
myotonica-containing WD repeat motif protein (DMR-N9
protein) (Protein 59) (Fragment), partial (14%)
Length = 1153
Score = 524 bits (1350), Expect = e-149
Identities = 258/286 (90%), Positives = 265/286 (92%), Gaps = 1/286 (0%)
Frame = +3
Query: 277 SFPILKDQTLFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDY 336
SFP++KDQT FSVAHARYSKSNPIARWHICQGSINSISFS DGAYLATVGRDGYLRVFDY
Sbjct: 3 SFPVVKDQTQFSVAHARYSKSNPIARWHICQGSINSISFSMDGAYLATVGRDGYLRVFDY 182
Query: 337 TKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSG 396
+KE L+CGGKSYYG LLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSG
Sbjct: 183 SKEQLICGGKSYYGALLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSG 362
Query: 397 VAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPLRRGPPGGSPTFGSP 456
VAFDSYWSSPNSNDNGETI YRFGSVGQDTQLLLW+LEMDEIVVPLRR PPG GSP
Sbjct: 363 VAFDSYWSSPNSNDNGETIMYRFGSVGQDTQLLLWDLEMDEIVVPLRR-PPG-----GSP 524
Query: 457 TFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACR 516
T+S GSQSSHWDN VPLGTLQPAPSM DVPKISPLVAHRVHTEPLS LIFTQESVLTACR
Sbjct: 525 TYSTGSQSSHWDNVVPLGTLQPAPSMLDVPKISPLVAHRVHTEPLSGLIFTQESVLTACR 704
Query: 517 EGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLSSKI-SNSIYKQ 561
EGHIKIW RPGVAESQ S SE LLAT+LKEKPSLSSKI SNS YKQ
Sbjct: 705 EGHIKIWMRPGVAESQSSTSENLLATNLKEKPSLSSKISSNSSYKQ 842
>TC226901 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia
myotonica-containing WD repeat motif protein (DMR-N9
protein) (Protein 59) (Fragment), partial (13%)
Length = 1242
Score = 515 bits (1326), Expect = e-146
Identities = 247/295 (83%), Positives = 266/295 (89%)
Frame = +3
Query: 267 EKNKDGAGESSFPILKDQTLFSVAHARYSKSNPIARWHICQGSINSISFSADGAYLATVG 326
EKN+DGAG+SSF ++KDQT FSV+HARYSKSNPIARWHICQGSI+SISFS DG Y+ATVG
Sbjct: 3 EKNRDGAGDSSFTVVKDQTQFSVSHARYSKSNPIARWHICQGSIDSISFSTDGTYIATVG 182
Query: 327 RDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAW 386
RDGYLRVFDY KE L+CGGKSYYG LLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAW
Sbjct: 183 RDGYLRVFDYLKEQLICGGKSYYGALLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAW 362
Query: 387 GEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWELEMDEIVVPLRRGP 446
GEGHNSWVSGVAFDSYWSSPNSNDN ET+ YRFGSVGQDTQLLLW+LEMDEIVVPLRR
Sbjct: 363 GEGHNSWVSGVAFDSYWSSPNSNDNEETVRYRFGSVGQDTQLLLWDLEMDEIVVPLRR-- 536
Query: 447 PGGSPTFGSPTFSAGSQSSHWDNAVPLGTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIF 506
P+ GSPT+SAGSQSS WD+ VPLGTLQPAPSM DV KISPLV HRVHTEPLS LIF
Sbjct: 537 ----PSGGSPTYSAGSQSSQWDSVVPLGTLQPAPSMLDVLKISPLVTHRVHTEPLSGLIF 704
Query: 507 TQESVLTACREGHIKIWTRPGVAESQPSNSETLLATSLKEKPSLSSKISNSIYKQ 561
TQESVLTACREGHIKIW RP AESQ SN+ETLL+TSLKEKPS+ SK+ NS ++Q
Sbjct: 705 TQESVLTACREGHIKIWIRPAAAESQSSNAETLLSTSLKEKPSILSKVGNSSHRQ 869
>TC230367
Length = 1152
Score = 379 bits (974), Expect = e-105
Identities = 193/242 (79%), Positives = 210/242 (86%), Gaps = 2/242 (0%)
Frame = +2
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T NG M + SSSS + +Q GLKTYFKTPEGRYKL ++KTHPSGLL + HGKTV
Sbjct: 446 TANGTMMSPSSSSVT-------AQSPGLKTYFKTPEGRYKLHYEKTHPSGLLHYAHGKTV 604
Query: 65 SMVTLAHLKEKPAPLTPTASSSS--FSASSGVRSAAARLLGGSNGNRALSFVGGNGSSKS 122
+ VTLAHLK+KPAP TPTASSSS FSASSGVRSAAARLLGGSNG+RALSFVGGNG SKS
Sbjct: 605 TQVTLAHLKDKPAPSTPTASSSSSSFSASSGVRSAAARLLGGSNGSRALSFVGGNGGSKS 784
Query: 123 NGGASRIGSIGSSSLSSSVANPNFDGKGSYLVFNAGDAILISDLNSQDKDPIKSIHFSNS 182
NGG +RIGSIG+SS SS++ANPNFDGKG+YL+FN GDAI ISDLNSQDKDPIKSIHFSNS
Sbjct: 785 NGGNTRIGSIGASSTSSAMANPNFDGKGTYLIFNVGDAIFISDLNSQDKDPIKSIHFSNS 964
Query: 183 NPVCHAFDQDAKDGHDLLIGLFSGDVYSVSLRQQLQDVGKKIVGAHHYNKDGILNNSRCT 242
NPVCHAFDQDAKDGHDLLIGL SGDVYSVSLRQQLQDVGKK V A HYN+DG +NNS CT
Sbjct: 965 NPVCHAFDQDAKDGHDLLIGLNSGDVYSVSLRQQLQDVGKKPVXALHYNRDGSVNNSHCT 1144
Query: 243 CI 244
CI
Sbjct: 1145CI 1150
>TC226900 similar to UP|DMWD_HUMAN (Q09019) Dystrophia myotonica-containing
WD repeat motif protein (DMR-N9 protein) (Protein 59)
(Fragment), partial (11%)
Length = 673
Score = 169 bits (428), Expect = 3e-42
Identities = 79/87 (90%), Positives = 82/87 (93%)
Frame = +2
Query: 355 CAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGET 414
C SMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGH+SWVSGVAFDSYWSSPNSNDN ET
Sbjct: 413 CFNSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHHSWVSGVAFDSYWSSPNSNDNEET 592
Query: 415 ITYRFGSVGQDTQLLLWELEMDEIVVP 441
+ YRFGSVGQDTQLLLW+L MDEIVVP
Sbjct: 593 VRYRFGSVGQDTQLLLWDL*MDEIVVP 673
Score = 63.2 bits (152), Expect = 3e-10
Identities = 25/29 (86%), Positives = 26/29 (89%)
Frame = +1
Query: 329 GYLRVFDYTKEHLVCGGKSYYGGLLCCAW 357
GYLRVFDY KE L+CGGKSYYG LLCCAW
Sbjct: 139 GYLRVFDYLKEQLICGGKSYYGALLCCAW 225
Score = 31.2 bits (69), Expect = 1.3
Identities = 15/28 (53%), Positives = 18/28 (63%)
Frame = +3
Query: 315 FSADGAYLATVGRDGYLRVFDYTKEHLV 342
FS DG Y+ATVGRDG +F H+V
Sbjct: 3 FSTDGTYIATVGRDG---IFFNPTHHIV 77
>BI974264
Length = 422
Score = 149 bits (375), Expect = 4e-36
Identities = 80/109 (73%), Positives = 88/109 (80%), Gaps = 1/109 (0%)
Frame = +1
Query: 5 TTNGNMTAASSSSSSGPTTTTQSQQQGLKTYFKTPEGRYKLQFDKTHPSGLLQFNHGKTV 64
T NG M + SSSS T +Q GLKTYFKT EGRYKLQ++KTHPSGLL + HGKTV
Sbjct: 112 TANGTMISPSSSSG------TAAQSPGLKTYFKTSEGRYKLQYEKTHPSGLLHYAHGKTV 273
Query: 65 SMVTLAHLKEKPAPLTPTA-SSSSFSASSGVRSAAARLLGGSNGNRALS 112
+ VTLAHLK+KPAP TPTA SSSSFSASSGVRSA ARLLGGSNG+RALS
Sbjct: 274 TQVTLAHLKDKPAPSTPTASSSSSFSASSGVRSAEARLLGGSNGSRALS 420
>AW277370
Length = 148
Score = 80.9 bits (198), Expect = 1e-15
Identities = 38/49 (77%), Positives = 45/49 (91%)
Frame = +2
Query: 474 GTLQPAPSMRDVPKISPLVAHRVHTEPLSSLIFTQESVLTACREGHIKI 522
GTLQPAPSM++VPKISP+V+HRVHTEPLS LIFTQES+LT ++ HIKI
Sbjct: 2 GTLQPAPSMQNVPKISPVVSHRVHTEPLSILIFTQESILTTYQKKHIKI 148
>TC209377 similar to UP|Q9FLX9 (Q9FLX9) Notchless protein homolog
(At5g52820), partial (33%)
Length = 669
Score = 53.1 bits (126), Expect = 3e-07
Identities = 37/138 (26%), Positives = 64/138 (45%)
Frame = +2
Query: 294 YSKSNPIARWHICQGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLL 353
+ +P R Q +N + FS DG ++A+ D +++++ T V + + G +
Sbjct: 86 FINKHPKTRMTGHQQLVNHVYFSPDGQWVASASFDKSVKLWNGTTGKFVTAFRGHVGPVY 265
Query: 354 CCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGE 413
+WS D + +L+G +D ++VW + RK+ GH V F WS +GE
Sbjct: 266 QISWSADSRLLLSGSKDSTLKVWDIRTRKLKQDLPGHADEV----FSVDWS-----PDGE 418
Query: 414 TITYRFGSVGQDTQLLLW 431
+ S G+D L LW
Sbjct: 419 KV----ASGGKDKVLKLW 460
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 48.9 bits (115), Expect = 6e-06
Identities = 24/94 (25%), Positives = 49/94 (51%)
Frame = +1
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S+ ++F DG+ A+ G D RV+D + + + +L ++S +G ++ TGG
Sbjct: 451 SVYGLAFHNDGSLAASCGLDSLARVWDLRTGRSILALEGHVKPVLGISFSPNGYHLATGG 630
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSY 402
ED+ ++W + +K H++ +S V F+ +
Sbjct: 631 EDNTCRIWDLRKKKSFYTIPAHSNLISQVKFEPH 732
Score = 39.7 bits (91), Expect = 0.004
Identities = 37/126 (29%), Positives = 53/126 (41%), Gaps = 7/126 (5%)
Frame = +1
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYY-----GGLLCCAW--SMDGKYIL 365
ISFS +G +LAT G D R++D K+ KS+Y L+ +G +++
Sbjct: 589 ISFSPNGYHLATGGEDNTCRIWDLRKK------KSFYTIPAHSNLISQVKFEPHEGYFLV 750
Query: 366 TGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQD 425
T D +VWS D K V GH + V+ V D G +T V D
Sbjct: 751 TASYDMTAKVWSGRDFKPVKTLSGHEAKVTSVDV--------LGDGGSIVT-----VSHD 891
Query: 426 TQLLLW 431
+ LW
Sbjct: 892 RTIKLW 909
Score = 32.3 bits (72), Expect = 0.58
Identities = 48/215 (22%), Positives = 85/215 (39%), Gaps = 3/215 (1%)
Frame = +1
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDL 372
+++S +LAT D + ++ + L+ + + L A+ GKY+ T D
Sbjct: 217 VAYSPVHDHLATASADRTAKYWN--QGSLLKTFEGHLDRLARIAFHPSGKYLGTASFDKT 390
Query: 373 VQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWE 432
++W +E + EGH+ V G+AF ND S G D+ +W+
Sbjct: 391 WRLWDIETGDELLLQEGHSRSVYGLAF--------HNDGSLA-----ASCGLDSLARVWD 531
Query: 433 LEMDEIVVPLRRGPPGGSPTFGSPTFSAGS---QSSHWDNAVPLGTLQPAPSMRDVPKIS 489
L ++ L P G +FS + DN + L+ S +P S
Sbjct: 532 LRTGRSILALEGHV---KPVLGI-SFSPNGYHLATGGEDNTCRIWDLRKKKSFYTIPAHS 699
Query: 490 PLVAHRVHTEPLSSLIFTQESVLTACREGHIKIWT 524
L++ +V EP ++TA + K+W+
Sbjct: 700 NLIS-QVKFEPHEGYF-----LVTASYDMTAKVWS 786
Score = 28.9 bits (63), Expect = 6.4
Identities = 14/45 (31%), Positives = 24/45 (53%)
Frame = +1
Query: 355 CAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAF 399
C++S DGK++ T ++WSM K + +GH + VA+
Sbjct: 91 CSFSRDGKWLATCSLTGASKLWSMPKIKKHSIFKGHTERATDVAY 225
>TC229097 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (47%)
Length = 755
Score = 47.8 bits (112), Expect = 1e-05
Identities = 30/130 (23%), Positives = 59/130 (45%), Gaps = 3/130 (2%)
Frame = +1
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM---DGKYILT 366
++ + FS + ++ D LR+++Y+ + + C + + +GKYI+
Sbjct: 136 VSFVKFSPNAKFILVGTLDNTLRLWNYSTGKFLKTYTGHVNSKYCISSTFSITNGKYIVG 315
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDT 426
G ED+ + +W ++ RK+V EGH+ V V+ P N G++G D
Sbjct: 316 GSEDNCIYLWDLQSRKIVQKLEGHSDAVVSVS-----CHPTEN------MIASGALGNDN 462
Query: 427 QLLLWELEMD 436
+ +W + D
Sbjct: 463 TVKIWTQQKD 492
>TC206061 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-like,
partial (36%)
Length = 657
Score = 47.4 bits (111), Expect = 2e-05
Identities = 24/80 (30%), Positives = 44/80 (55%)
Frame = +3
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
Q ++S+S+S + L T G + +R +D + + + GL+ CAW GKYIL+
Sbjct: 345 QKPVSSVSWSPNDQELLTCGVEEAVRRWDVSTGTCLQVYEKNGPGLISCAWFPSGKYILS 524
Query: 367 GGEDDLVQVWSMEDRKVVAW 386
G D + +W ++ ++V +W
Sbjct: 525 GLSDKSICMWDLDGKEVESW 584
>TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%)
Length = 1048
Score = 47.0 bits (110), Expect = 2e-05
Identities = 26/92 (28%), Positives = 46/92 (49%)
Frame = +2
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGE 369
++ + + A+ Y+AT D +R++D V + G +L A S DG+Y+ +G E
Sbjct: 248 VDCVQWHANCNYIATGSSDKTVRLWDVQSGECVRVFVGHRGMILSLAMSPDGRYMASGDE 427
Query: 370 DDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS 401
D + +W + + + GH S V +AF S
Sbjct: 428 DGTIMMWDLSSGRCLTPLIGHTSCVWSLAFSS 523
Score = 42.0 bits (97), Expect = 7e-04
Identities = 28/130 (21%), Positives = 51/130 (38%)
Frame = +2
Query: 313 ISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDDL 372
+ FS G Y A+ D R++ + + + + C W + YI TG D
Sbjct: 131 VQFSPVGHYFASSSHDRTARIWSMDRIQPLRIMAGHLSDVDCVQWHANCNYIATGSSDKT 310
Query: 373 VQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQLLLWE 432
V++W ++ + V GH + +A SP+ S +D +++W+
Sbjct: 311 VRLWDVQSGECVRVFVGHRGMILSLAM-----SPDGR--------YMASGDEDGTIMMWD 451
Query: 433 LEMDEIVVPL 442
L + PL
Sbjct: 452 LSSGRCLTPL 481
>TC206060 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-like,
partial (62%)
Length = 1510
Score = 45.1 bits (105), Expect = 9e-05
Identities = 23/80 (28%), Positives = 43/80 (53%)
Frame = +3
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
Q ++S+S+S + L T G + +R +D + + + GL+ C+W GKYIL
Sbjct: 558 QKPVSSVSWSPNDQELLTCGVEEAIRRWDVSTGKCLQIYEKAGAGLVSCSWFPCGKYILC 737
Query: 367 GGEDDLVQVWSMEDRKVVAW 386
G D + +W ++ ++V +W
Sbjct: 738 GLSDKSICMWELDGKEVESW 797
>TC207724 similar to PIR|A96839|A96839 F23A5.2(form2) [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(98%)
Length = 1343
Score = 44.7 bits (104), Expect = 1e-04
Identities = 38/139 (27%), Positives = 60/139 (42%), Gaps = 24/139 (17%)
Frame = +2
Query: 295 SKSNPIARWHICQ---GSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGK----- 346
+ +NP + + Q SI+SI FS +L D +R ++ T+ V
Sbjct: 92 ANTNPNKSYEVAQPPSDSISSICFSPKANFLVATSWDNQVRCWEITRNGTVVNSTPKASI 271
Query: 347 SYYGGLLCCAWSMDGKYILTGGEDDLVQVW-----------SMEDRKV--VAWGEGHNSW 393
S+ +LC AW DG + +GG D V++W +M D V +AW N
Sbjct: 272 SHDQPVLCSAWKDDGTTVFSGGCDKQVKMWPLMSGGQPMTVAMHDAPVKDIAWIPEMNLL 451
Query: 394 VSGVAFD---SYWSSPNSN 409
+G ++D YW + SN
Sbjct: 452 ATG-SWDKTLKYWDTRQSN 505
>TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (57%)
Length = 1124
Score = 44.7 bits (104), Expect = 1e-04
Identities = 33/138 (23%), Positives = 62/138 (44%), Gaps = 5/138 (3%)
Frame = +1
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGG-----KSYYGGLLCCAWSMDGKYI 364
I+ +++S+D Y+ + D LR++D T V GG + + + C ++ YI
Sbjct: 271 ISDLAWSSDSHYICSASDDRTLRIWDAT----VGGGCIKILRGHDDAVFCVNFNPQSSYI 438
Query: 365 LTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQ 424
++G D+ ++VW ++ K V +GH V+ V + N +G I S
Sbjct: 439 VSGSFDETIKVWDVKTGKCVHTIKGHTMPVTSVHY---------NRDGNLII----SASH 579
Query: 425 DTQLLLWELEMDEIVVPL 442
D +W+ E ++ L
Sbjct: 580 DGSCKIWDTETGNLLKTL 633
Score = 36.2 bits (82), Expect = 0.040
Identities = 30/114 (26%), Positives = 47/114 (40%), Gaps = 5/114 (4%)
Frame = +1
Query: 307 QGSINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILT 366
+ +++ + FS DG LA+ D L ++ L + G+ AWS D YI +
Sbjct: 136 ENAVSCVKFSNDGTLLASASLDKTLIIWSSATLTLCHRLVGHSEGISDLAWSSDSHYICS 315
Query: 367 GGEDDLVQVWSMEDRKVVAWG-----EGHNSWVSGVAFDSYWSSPNSNDNGETI 415
+D +++W V G GH+ V V F+ S S ETI
Sbjct: 316 ASDDRTLRIWD----ATVGGGCIKILRGHDDAVFCVNFNPQSSYIVSGSFDETI 465
Score = 32.3 bits (72), Expect = 0.58
Identities = 16/38 (42%), Positives = 23/38 (60%)
Frame = +2
Query: 360 DGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGV 397
+GKYI+ G ED V +W ++ +K+V EGH V V
Sbjct: 815 NGKYIVGGSEDHCVYIWDLQ-QKLVQKLEGHTDTVISV 925
>TC231248
Length = 849
Score = 44.3 bits (103), Expect = 1e-04
Identities = 33/105 (31%), Positives = 51/105 (48%), Gaps = 6/105 (5%)
Frame = +1
Query: 304 HICQGSINSISFSADGAYLATVGR-DGYLRVFDYTKEHLVCGGKSYYGG-----LLCCAW 357
H +G I + FS DG YL T GR D Y+ +D K + C K Y +
Sbjct: 148 HGQEGGITHVQFSRDGNYLYTGGRKDPYILCWDVRKS-VDCVYKLYRSSENTNQRILFDI 324
Query: 358 SMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSY 402
GKY+ TGG+D LV +++++ + V+ E V+G +F +
Sbjct: 325 DPSGKYLGTGGQDGLVHIYNLQTGQWVSSFEAALDTVNGFSFHPF 459
>TC228105 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;} , complete
Length = 1113
Score = 43.1 bits (100), Expect = 3e-04
Identities = 35/124 (28%), Positives = 53/124 (42%), Gaps = 1/124 (0%)
Frame = +3
Query: 310 INSISFSA-DGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
+ S+ +S D L T DG + ++D + L+ + +LC S DG I TG
Sbjct: 603 VRSLVYSPYDPRLLFTASDDGNVHMYDAEGKALIGTMSGHASWVLCVDVSPDGAAIATGS 782
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITYRFGSVGQDTQL 428
D V++W + R V H+ V GVAF P +D R SV D +
Sbjct: 783 SDRSVRLWDLNMRASVQTMSNHSDQVWGVAF----RPPGGSD---VRGGRLASVSDDKSI 941
Query: 429 LLWE 432
L++
Sbjct: 942 SLYD 953
Score = 31.2 bits (69), Expect = 1.3
Identities = 16/51 (31%), Positives = 24/51 (46%)
Frame = +3
Query: 352 LLCCAWSMDGKYILTGGEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSY 402
+L AWS DGK + G D + V+ + K + EGH V + + Y
Sbjct: 477 VLSVAWSPDGKRLACGSMDGTISVFDVPRAKFLHHLEGHFMPVRSLVYSPY 629
>TC228062 weakly similar to UP|Q39221 (Q39221) St12p protein, partial (54%)
Length = 1534
Score = 42.7 bits (99), Expect = 4e-04
Identities = 32/124 (25%), Positives = 54/124 (42%), Gaps = 1/124 (0%)
Frame = +3
Query: 312 SISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGGEDD 371
+++F+ DG LA G DG LRVF + ++ + + L +S DGK + + G
Sbjct: 507 ALAFNNDGTALAAGGEDGNLRVFKWPSMEIILNETNAHSSLKDLHFSSDGKLLASLGSGG 686
Query: 372 LVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGET-ITYRFGSVGQDTQLLL 430
+VW + V+ S +S +++ S S N ET I Y + +L
Sbjct: 687 PCKVWDVSSSMVL-------SSLSNENRETFSSCRFSQTNDETLILYIAAMTDKGGSILT 845
Query: 431 WELE 434
W +
Sbjct: 846 WNTQ 857
>TC221163
Length = 832
Score = 42.4 bits (98), Expect = 6e-04
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Frame = +3
Query: 309 SINSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGK-SYYGGLLCCAWSMDGKYILTG 367
SI + F+ G++ A G D L ++D EHL+ G + G+ C AW + Y+ +G
Sbjct: 342 SIECVGFAPSGSWAAVGGMDKKLIIWDI--EHLLPRGTCEHEDGVTCLAW-LGASYVASG 512
Query: 368 GEDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDS 401
D V++W + V +GH+ + ++ S
Sbjct: 513 CVDGKVRLWDSRSGECVKTLKGHSDAIQSLSVSS 614
>TC206579 weakly similar to UP|TAF5_YEAST (P38129) Transcription initiation
factor TFIID subunit 5 (TBP-associated factor 5)
(TBP-associated factor 90 kDa) (TAFII-90), partial (4%)
Length = 835
Score = 42.0 bits (97), Expect = 7e-04
Identities = 27/109 (24%), Positives = 48/109 (43%), Gaps = 3/109 (2%)
Frame = +1
Query: 312 SISFSADGAYLATVGRDGYLRVFD---YTKEHLVCGGKSYYGGLLCCAWSMDGKYILTGG 368
S F YLAT D +++++ +T E + G + + C +S+DG Y++T
Sbjct: 313 SPEFCEPHRYLATASSDHTVKIWNVDGFTLEKTLIGHQRWVWD---CVFSVDGAYLITAS 483
Query: 369 EDDLVQVWSMEDRKVVAWGEGHNSWVSGVAFDSYWSSPNSNDNGETITY 417
D ++WSM + + +GH+ A S DNG + +
Sbjct: 484 SDTTARLWSMSTGEDIKVYQGHHKATICCALHDGAEPATS*DNGHKVIF 630
>TC229098 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (59%)
Length = 580
Score = 41.6 bits (96), Expect = 0.001
Identities = 20/85 (23%), Positives = 44/85 (51%), Gaps = 3/85 (3%)
Frame = +3
Query: 310 INSISFSADGAYLATVGRDGYLRVFDYTKEHLVCGGKSYYGGLLCCAWSM---DGKYILT 366
++ + FS + ++ D LR+++Y+ + + C + + +GKYI+
Sbjct: 318 VSFVKFSPNAKFILVGTLDNTLRLWNYSTGKFLKTYTGHVNSKYCISSTFSTTNGKYIVG 497
Query: 367 GGEDDLVQVWSMEDRKVVAWGEGHN 391
G E++ + +W ++ RK+V EGH+
Sbjct: 498 GSEENYIYLWDLQSRKIVQKLEGHS 572
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,481,003
Number of Sequences: 63676
Number of extensions: 443204
Number of successful extensions: 2685
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 2374
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2572
length of query: 561
length of database: 12,639,632
effective HSP length: 102
effective length of query: 459
effective length of database: 6,144,680
effective search space: 2820408120
effective search space used: 2820408120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC128638.4


