

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.4 + phase: 0
(301 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
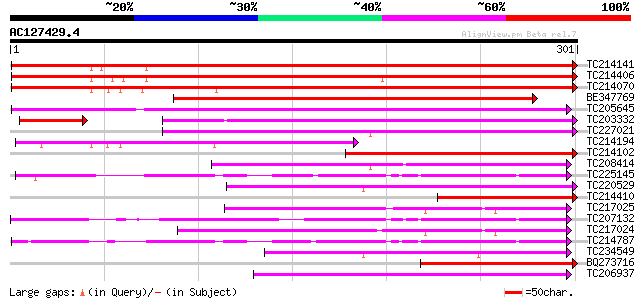
Score E
Sequences producing significant alignments: (bits) Value
TC214141 similar to UP|Q947D4 (Q947D4) Dehydration-responsive pr... 354 2e-98
TC214406 weakly similar to UP|Q947D4 (Q947D4) Dehydration-respon... 347 5e-96
TC214070 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced prote... 346 6e-96
BE347769 weakly similar to GP|16588826|gb dehydration-responsive... 214 5e-56
TC205645 UP|Q8RUD9 (Q8RUD9) Seed coat BURP domain protein 1, com... 205 2e-53
TC203332 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific... 198 2e-51
TC227021 weakly similar to UP|O65009 (O65009) BURP domain contai... 176 9e-45
TC214194 weakly similar to UP|Q94IC5 (Q94IC5) BURP domain-contai... 175 2e-44
TC214102 similar to UP|Q9MB24 (Q9MB24) S1-3 protein (Fragment), ... 171 4e-43
TC208414 weakly similar to UP|O65009 (O65009) BURP domain contai... 155 2e-38
TC225145 homologue to UP|O24482 (O24482) Sali3-2, complete 135 2e-32
TC220529 weakly similar to GB|CAE02618.1|38567637|OSA575668 RAFT... 134 5e-32
TC214410 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced prote... 123 9e-29
TC217025 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzy... 121 3e-28
TC207132 similar to UP|EA92_VICFA (P21747) Embryonic abundant pr... 120 1e-27
TC217024 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzy... 119 1e-27
TC214787 UP|Q06765 (Q06765) ADR6 protein (Sali5-4a protein), com... 117 5e-27
TC234549 weakly similar to UP|O65009 (O65009) BURP domain contai... 109 1e-24
BQ273716 weakly similar to SP|Q08298|RD22_ Dehydration-responsiv... 106 1e-23
TC206937 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzy... 102 2e-22
>TC214141 similar to UP|Q947D4 (Q947D4) Dehydration-responsive protein RD22
(Fragment), partial (89%)
Length = 1353
Score = 354 bits (909), Expect = 2e-98
Identities = 182/334 (54%), Positives = 229/334 (68%), Gaps = 34/334 (10%)
Frame = +1
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP------------------M 43
I +L +ALVATHA LPPE+YWKS LPTT +PKAITDIL+ +
Sbjct: 97 IFTLLNIALVATHAALPPEVYWKSVLPTTPMPKAITDILYSDWVEEKSSSVHVGGGGVNV 276
Query: 44 DDGK-------ISVDGKISVNGKTRIVYDAAPIYF---------YENDANEAELHDNRNL 87
GK ++V GK + P++ Y A E +LHD+ N+
Sbjct: 277 HTGKGGGSGTTVNVGGKGGGGVNVHAGHKGKPVHVSVGSKSPFDYVYAATETQLHDDPNV 456
Query: 88 AMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTE 147
A+F LEKDLH GTK ++ FT+++ + TFL VA+SIPFSSNK++ I N FS+K GS E
Sbjct: 457 ALFFLEKDLHSGTKLDLHFTRSTSNQATFLSRQVADSIPFSSNKVDFIFNKFSVKPGSEE 636
Query: 148 SEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVI 207
++I+KNTISECE GIKGEEK C TSLESM+DF+T KLGNNV+ VSTEV+ E+GLQ+Y +
Sbjct: 637 AQIMKNTISECEEGGIKGEEKYCATSLESMVDFSTSKLGNNVEVVSTEVDKETGLQKYTV 816
Query: 208 ANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQ 267
A GVKK+ + VVCHK+NYPYAVFYCHKT+ T+ YSVPLEG +G RVKAVA+CH+DTS+
Sbjct: 817 APGVKKLSGDKAVVCHKQNYPYAVFYCHKTETTRAYSVPLEGTNGVRVKAVAVCHTDTSE 996
Query: 268 WSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
W+ KHLAFQVLKV+ GT PVCH L + VVW K
Sbjct: 997 WNPKHLAFQVLKVKPGTIPVCHFLPEDHVVWVPK 1098
>TC214406 weakly similar to UP|Q947D4 (Q947D4) Dehydration-responsive protein
RD22 (Fragment), partial (90%)
Length = 1379
Score = 347 bits (889), Expect = 5e-96
Identities = 183/336 (54%), Positives = 229/336 (67%), Gaps = 36/336 (10%)
Frame = +1
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHP----MDDGKISVDGK---- 53
I +L LALVA HA LPPE+YWKS LPTT +PKAITDIL+P ++V GK
Sbjct: 145 IFTLLNLALVAIHAALPPEVYWKSVLPTTPMPKAITDILYPDWVEEKSTSVNVGGKGVNV 324
Query: 54 ----------ISVNGK-------TRIVYDAAPIYF---------YENDANEAELHDNRNL 87
++V GK + P++ Y + E +LHD+ N+
Sbjct: 325 HAGKGGGGTNVNVGGKGSGGGVNVHAGHKGKPVHVSVGSKSPFNYIYASTETQLHDDPNV 504
Query: 88 AMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTE 147
A+F LEKDLH GTK N+ FT +S+ TFLP VA+SIPFSS+K+E + N FS+K GS E
Sbjct: 505 ALFFLEKDLHPGTKLNLHFTTSSNIQATFLPRQVADSIPFSSSKVEVVFNKFSVKPGSEE 684
Query: 148 SEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEV--NGESGLQQY 205
++I+KNT+SECE GIKGEEK C TSLESMIDF+T KLG NV+ VSTEV + E+GLQ+Y
Sbjct: 685 AQIMKNTLSECEEGGIKGEEKYCATSLESMIDFSTSKLGKNVEVVSTEVVEDKETGLQKY 864
Query: 206 VIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDT 265
+A GV K+ + VVCHK+NYPYAVFYCHKT+ T+ YSVPLEGA+G RVKAVA+CH+ T
Sbjct: 865 TVAPGVNKLSGDKAVVCHKQNYPYAVFYCHKTETTRAYSVPLEGANGVRVKAVAVCHTHT 1044
Query: 266 SQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
S+W+ KHLAFQVLKV+ GT PVCH L + VVW K
Sbjct: 1045SEWNPKHLAFQVLKVKPGTVPVCHFLPEDHVVWVPK 1152
>TC214070 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced protein RD22-like
protein, partial (81%)
Length = 1427
Score = 346 bits (888), Expect = 6e-96
Identities = 196/360 (54%), Positives = 238/360 (65%), Gaps = 60/360 (16%)
Frame = +3
Query: 2 INVVLQLALVATHA-TLPPELYWKSKLPTTQIPKAITDILHP-----------MDDGKIS 49
I +L LA+VATHA TLPPE+YWKSKLPTT +PKAITDILHP + G ++
Sbjct: 117 IFALLNLAVVATHAETLPPEVYWKSKLPTTPMPKAITDILHPDLAEDKSTSVAVGKGGVN 296
Query: 50 VD---------------GKISVN--------------GKTRIVYDAAPI----------- 69
V+ G ++VN GK + + P
Sbjct: 297 VNAGKTKPGGTSVNVGKGGVNVNTGKGKPNKGTSVNVGKGGVNVNTGPKKGKPVHVGVGP 476
Query: 70 ---YFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTK--TSDHGPTFLPGDVANS 124
+ Y A+E + HD+ N+A+F LEKDLH+GTK N+ FT+ TS +FLP VA+S
Sbjct: 477 HSPFDYNYAASETQWHDDPNVALFFLEKDLHYGTKLNLHFTRYFTSSVDASFLPRSVADS 656
Query: 125 IPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLK 184
IPFSSNK+ +LN FSIK+GS E++ VKNTISECE GIKGEEK CVTSLESM+DF T K
Sbjct: 657 IPFSSNKVNEVLNKFSIKEGSDEAQTVKNTISECEVPGIKGEEKRCVTSLESMVDFATTK 836
Query: 185 LGNN-VDTVSTEVNGESG-LQQYVIANGVKKMGENNL-VVCHKRNYPYAVFYCHKTDATK 241
LG+N VD VSTEV + LQQY +A GVK++GE+ VVCHK NYPYAVFYCHK++ TK
Sbjct: 837 LGSNDVDAVSTEVTKKDNELQQYTMAPGVKRLGEDKASVVCHKENYPYAVFYCHKSETTK 1016
Query: 242 VYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
YSVPLEGADGSRVKAVA+CH+DTS+W+ KHLAFQVLKVQ GT PVCH L Q VV+ K
Sbjct: 1017AYSVPLEGADGSRVKAVAVCHTDTSKWNPKHLAFQVLKVQPGTVPVCHFLPQDHVVFVPK 1196
>BE347769 weakly similar to GP|16588826|gb dehydration-responsive protein
RD22 {Prunus persica}, partial (38%)
Length = 595
Score = 214 bits (544), Expect = 5e-56
Identities = 110/193 (56%), Positives = 140/193 (71%)
Frame = +2
Query: 88 AMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTE 147
A++L+E DLH GTK ++ FT+++ + TFL VA+SIPFSSNK++ I N S K GS E
Sbjct: 2 ALYLVEFDLHSGTKLDLHFTRSTSNQATFLSRQVADSIPFSSNKVDFIFNKNSGKPGSKE 181
Query: 148 SEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVI 207
++I+KNTI ECE GIKGEEK C TSLESM DF+T KLGNNV+ VSTEV+ ++G +Y
Sbjct: 182 AQIMKNTIIECEEGGIKGEEKYCATSLESMDDFSTSKLGNNVEVVSTEVDKDTG*HKYTG 361
Query: 208 ANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQ 267
A GVKK+ + VVCHK NYPYA FYC++TD T YSVP EG + R KAV+ CH+DTS
Sbjct: 362 APGVKKVSGDKAVVCHKHNYPYADFYCNRTDTTIAYSVPSEGTNVVRDKAVSRCHTDTSD 541
Query: 268 WSLKHLAFQVLKV 280
+H+A VL V
Sbjct: 542 MYPRHVANHVLTV 580
>TC205645 UP|Q8RUD9 (Q8RUD9) Seed coat BURP domain protein 1, complete
Length = 1369
Score = 205 bits (521), Expect = 2e-53
Identities = 109/299 (36%), Positives = 163/299 (54%), Gaps = 2/299 (0%)
Frame = +1
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTR 61
I + L L L+ +A L P YW++ LP T +PKAIT++L G +
Sbjct: 58 IFLYLNLMLMTANAALTPRHYWETMLPRTPLPKAITELLSLESRSIFEYAGNDDQSESRS 237
Query: 62 IVYDAAPIYFYENDANEAELHDNRNL-AMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGD 120
I+ A Y D ++ H+ + +F LE+DL G FN++F + LP
Sbjct: 238 ILGYAG----YNQDEDDVSKHNIQIFNRLFFLEEDLRAGKIFNMKFVNNTKATVPLLPRQ 405
Query: 121 VANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDF 180
++ IPFS +K + +L ++ S+ ++I+ TI C+ +GE K C TSLESM+DF
Sbjct: 406 ISKQIPFSEDKKKQVLAMLGVEANSSNAKIIAETIGLCQEPATEGERKHCATSLESMVDF 585
Query: 181 TTLKLGNNVDTVSTEVNGESGLQQYVIA-NGVKKMGENNLVVCHKRNYPYAVFYCHKTDA 239
LG NV STE E+ ++V+ NGV+K+G++ ++ CH +YPY VF CH
Sbjct: 586 VVSALGKNVGAFSTEKERETESGKFVVVKNGVRKLGDDKVIACHPMSYPYVVFGCHLVPR 765
Query: 240 TKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ Y V L+G DG RVKAV CH DTS+W H AF+VL ++ G VCH+ +G ++W
Sbjct: 766 SSGYLVRLKGEDGVRVKAVVACHRDTSKWDHNHGAFKVLNLKPGNGTVCHVFTEGNLLW 942
>TC203332 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein-2,
partial (60%)
Length = 1444
Score = 198 bits (504), Expect = 2e-51
Identities = 95/220 (43%), Positives = 131/220 (59%)
Frame = +1
Query: 82 HDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSI 141
HD E+ L GTK + F K + P LP +A IP SS K++ I+ +
Sbjct: 583 HDIPKADQVFFEEGLRPGTKLDAHFKKRENVTP-LLPRQIAQHIPLSSAKIKEIVEMLFV 759
Query: 142 KQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESG 201
+I++ TIS CE I GEE+ C TSLESM+DF T KLG N +STE ES
Sbjct: 760 NPEPENVKILEETISMCEVPAITGEERYCATSLESMVDFVTSKLGKNARVISTEAEKESK 939
Query: 202 LQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAIC 261
Q++ + +GVK + E+ ++VCH +YPY VF CH+ T + +PLEG DG+RVKA A+C
Sbjct: 940 SQKFSVKDGVKLLAEDKVIVCHPMDYPYVVFMCHEISNTTAHFMPLEGEDGTRVKAAAVC 1119
Query: 262 HSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
H DTS+W H+ Q+LK + G PVCHI +G ++WF+K
Sbjct: 1120HKDTSEWDPNHVFLQMLKTKPGAAPVCHIFPEGHLLWFAK 1239
Score = 46.6 bits (109), Expect = 1e-05
Identities = 19/36 (52%), Positives = 25/36 (68%)
Frame = +1
Query: 6 LQLALVATHATLPPELYWKSKLPTTQIPKAITDILH 41
L L L+ HA +PPE+YW+ LP T +PKAI D L+
Sbjct: 46 LNLLLMTAHAAIPPEVYWERMLPNTPMPKAIIDFLN 153
>TC227021 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (47%)
Length = 1147
Score = 176 bits (447), Expect = 9e-45
Identities = 91/226 (40%), Positives = 128/226 (56%), Gaps = 6/226 (2%)
Frame = +1
Query: 82 HDNRNLAMFLLEKDLHHGTKFNIQFTKTSDH-GPTFLPGDVANSIPFSSNKLENILNYFS 140
H + ++ +F +DL G I F K P P + A+S+PFS NKL N+L FS
Sbjct: 295 HIDPSVMVFFTIEDLKVGKTMPIHFPKRDPATSPKLWPREEADSLPFSLNKLPNLLKIFS 474
Query: 141 IKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVD----TVSTEV 196
+ Q S +++ +++T+ ECE IKGE K C TSLESM+DFT LG D + S +
Sbjct: 475 VSQNSPKAKAMEDTLRECETKPIKGEVKFCATSLESMLDFTQSILGFTSDLKVLSTSHQT 654
Query: 197 NGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDA-TKVYSVPLEGADGSRV 255
Q Y + + ++ + +V CH YPY VFYCH ++ K+Y VPL G +G RV
Sbjct: 655 KSSVTFQNYTMLENIIEIPASKMVACHTMPYPYTVFYCHSQESENKIYRVPLAGENGDRV 834
Query: 256 KAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVWFSK 301
A+ +CH DTSQW H++FQVLKV+ GT VCH ++W K
Sbjct: 835 DAMVVCHMDTSQWGHGHVSFQVLKVKPGTTSVCHFFPADHLIWVPK 972
>TC214194 weakly similar to UP|Q94IC5 (Q94IC5) BURP domain-containing protein
(Fragment), partial (49%)
Length = 704
Score = 175 bits (444), Expect = 2e-44
Identities = 106/222 (47%), Positives = 134/222 (59%), Gaps = 40/222 (18%)
Frame = +1
Query: 4 VVLQLALVATHA--TLPPELYWKSKLPTTQIPKAITDILHP---------MDDGKISV-- 50
+ + +VAT+A LPP YWKSKLPTT +PKAITD+L P +D GK V
Sbjct: 37 ITFLMLVVATNADAALPPAFYWKSKLPTTPMPKAITDLLQPDWKEEKDTSVDVGKGGVNV 216
Query: 51 --------------------DGKISVN----GK-TRIVYDAAPIYFYENDANEAELHDNR 85
DG ++V+ GK + + Y A+E +LHD+
Sbjct: 217 GVEKGNQGPGDGTDVNVGGGDGGVNVHTGPKGKPVHVGVGPHSPFDYNYAASETQLHDDP 396
Query: 86 NLAMFLLEKDLHHGTKFNIQFT--KTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQ 143
N+A+F LEKDLHHGTK N+ FT TS+ TFLP V++SIPFSSNK+ ++LN FSIK
Sbjct: 397 NVALFFLEKDLHHGTKLNLHFTIYYTSNVDATFLPRSVSDSIPFSSNKVNDVLNKFSIKD 576
Query: 144 GSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKL 185
GS E++ VKNTI+ECE IKGEEK CVTSLESM+DF T KL
Sbjct: 577 GSDEAKTVKNTINECEGPSIKGEEKRCVTSLESMVDFATTKL 702
>TC214102 similar to UP|Q9MB24 (Q9MB24) S1-3 protein (Fragment), partial
(70%)
Length = 621
Score = 171 bits (433), Expect = 4e-43
Identities = 85/126 (67%), Positives = 99/126 (78%), Gaps = 3/126 (2%)
Frame = +3
Query: 179 DFTTLKLGNN-VDTVSTEVNGESG-LQQYVIANGVKKMGENNL-VVCHKRNYPYAVFYCH 235
DF T KLG+N VD VSTEV + LQQY +A GVK++GE+ VVCHK NYPYAVFYCH
Sbjct: 15 DFATTKLGSNDVDAVSTEVTKKDNELQQYTMAPGVKRLGEDKASVVCHKENYPYAVFYCH 194
Query: 236 KTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQ 295
K++ TK YSVPLEGADGSRVKAVA+CH+DTS+W+ KHLAFQVLKV GT P+CH L Q
Sbjct: 195 KSETTKAYSVPLEGADGSRVKAVAVCHTDTSKWNPKHLAFQVLKVHPGTVPICHFLPQDH 374
Query: 296 VVWFSK 301
VV+ K
Sbjct: 375 VVFVPK 392
>TC208414 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (39%)
Length = 957
Score = 155 bits (393), Expect = 2e-38
Identities = 83/196 (42%), Positives = 112/196 (56%), Gaps = 5/196 (2%)
Frame = +2
Query: 108 KTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEE 167
KT H F P FSS L ++L +FSI + S +++ +K T+ +CE ++GE
Sbjct: 215 KTPQHLQNF*PEKKLTRFLFSSKHLPSLLKFFSIPKHSPQAKAMKYTLKQCEFEPMEGET 394
Query: 168 KLCVTSLESMIDFTTLKLGNNVD----TVSTEVNGESGLQQYVIANGVKKMGENNLVVCH 223
K C TSLES+ DF G+N T N + LQ Y I+ VK + N++ CH
Sbjct: 395 KFCATSLESLFDFAHYLFGSNAQFKVLTTVHLTNSTTLLQNYTISE-VKVISVPNVIGCH 571
Query: 224 KRNYPYAVFYCHKTDA-TKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQ 282
YPYAVFYCH + T +Y V +EG +G RV+A AICH DTS+W H++F+VLKVQ
Sbjct: 572 PMPYPYAVFYCHSQHSDTNLYEVMVEGENGGRVQAAAICHMDTSKWDRDHVSFRVLKVQP 751
Query: 283 GTFPVCHILQQGQVVW 298
GT PVCH +VW
Sbjct: 752 GTSPVCHFFPPDNLVW 799
>TC225145 homologue to UP|O24482 (O24482) Sali3-2, complete
Length = 1382
Score = 135 bits (340), Expect = 2e-32
Identities = 93/298 (31%), Positives = 139/298 (46%), Gaps = 3/298 (1%)
Frame = +3
Query: 4 VVLQLALVA---THATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKT 60
++ LAL HA+LP E YW++ P T IP A+ ++L P+ G
Sbjct: 207 ILFSLALAGESHVHASLPEEDYWEAVWPNTPIPTALRELLKPLPAG-------------- 344
Query: 61 RIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGD 120
E D ++ D + F ++DLH G +QFTK P P
Sbjct: 345 -----------VEIDELPKQIDDTQYPKTFFYKEDLHPGKTMKVQFTKR----PYAQPYG 479
Query: 121 VANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDF 180
V + IK S E + + EA+ +GEEK C SL ++I F
Sbjct: 480 VYTWLT-------------DIKDTSKEGYSFEEICIKKEAF--EGEEKFCAKSLGTVIGF 614
Query: 181 TTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDAT 240
KLG N+ +S+ + +QY + GV+ +G+ V+CH N+ AVFYCHK T
Sbjct: 615 AISKLGKNIQVLSSSFVNKQ--EQYTV-EGVQNLGDK-AVMCHGLNFRTAVFYCHKVRET 782
Query: 241 KVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ VPL DG++ +A+A+CHSDTS + H+ +++ V GT PVCH L ++W
Sbjct: 783 TAFMVPLVAGDGTKTQALAVCHSDTSGMN-HHMLHELMGVDPGTNPVCHFLGSKAILW 953
>TC220529 weakly similar to GB|CAE02618.1|38567637|OSA575668 RAFTIN1 protein
{Oryza sativa (japonica cultivar-group);} , partial
(15%)
Length = 942
Score = 134 bits (337), Expect = 5e-32
Identities = 71/192 (36%), Positives = 108/192 (55%), Gaps = 6/192 (3%)
Frame = +3
Query: 116 FLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLE 175
FLP A SIPFS ++L ++L FSI + S E+ +++T+ +CEA I GE K+C TSLE
Sbjct: 216 FLPRKEAESIPFSISQLPSVLQLFSISEDSPEANAMRDTLEQCEAEPITGETKICATSLE 395
Query: 176 SMIDFTTLKLG----NNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAV 231
SM++F +G +N+ T + LQ++ I + + V CH YPYA+
Sbjct: 396 SMLEFIGTIIGSETKHNILTTTLPTASGVPLQKFTILEVSEDINAAKWVACHPLPYPYAI 575
Query: 232 FYCH-KTDATKVYSVPLEGADG-SRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCH 289
+YCH +KV+ V L +G +++A+ ICH DTS WS H+ F+ L ++ G VCH
Sbjct: 576 YYCHFIATGSKVFKVSLGSENGDDKIEALGICHLDTSDWSPNHIIFRQLGIKPGKDAVCH 755
Query: 290 ILQQGQVVWFSK 301
++W K
Sbjct: 756 FFPIKHLMWVPK 791
>TC214410 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced protein RD22-like
protein, partial (22%)
Length = 575
Score = 123 bits (309), Expect = 9e-29
Identities = 56/74 (75%), Positives = 63/74 (84%)
Frame = +1
Query: 228 PYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPV 287
PYAVFYCHK++ TK YSVPLEGADGSRVKAVA+CH+DTS+W+ KHLAFQVLKVQ GT PV
Sbjct: 121 PYAVFYCHKSETTKAYSVPLEGADGSRVKAVAVCHTDTSKWNPKHLAFQVLKVQPGTVPV 300
Query: 288 CHILQQGQVVWFSK 301
CH L Q VV+ K
Sbjct: 301 CHFLPQDHVVFVPK 342
>TC217025 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzyme 1 beta
subunit precursor , partial (32%)
Length = 933
Score = 121 bits (304), Expect = 3e-28
Identities = 69/191 (36%), Positives = 101/191 (52%), Gaps = 7/191 (3%)
Frame = +2
Query: 115 TFLPGDVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSL 174
+FLP + +PFSS+K+ + F + S+ +++ +++ ECE GE K CV S+
Sbjct: 41 SFLPRSILTKLPFSSSKVHELKRLFKVSDNSSMEKMIMDSLGECERVPSMGETKRCVGSI 220
Query: 175 ESMIDFTTLKLGNNVDTVSTE-VNGESGLQQYVIANGVKKMGENNL---VVCHKRNYPYA 230
E MIDF+T LG NV +TE VNG + + V+ VK M + V CH+ +PY
Sbjct: 221 EDMIDFSTSVLGRNVAVWTTENVNGSN---KNVMVGRVKGMNGGKVTQSVSCHQSLFPYM 391
Query: 231 VFYCHKTDATKVYSVPLEGADGSRVK---AVAICHSDTSQWSLKHLAFQVLKVQQGTFPV 287
++YCH +VY L + S+ K VAICH DT+ WS H AF L G V
Sbjct: 392 LYYCHSVPKVRVYQADLLDPE-SKAKINHGVAICHLDTTAWSPTHGAFMALGSGPGRIEV 568
Query: 288 CHILQQGQVVW 298
CH + + + W
Sbjct: 569 CHWIFENDLTW 601
>TC207132 similar to UP|EA92_VICFA (P21747) Embryonic abundant protein USP92
precursor, partial (51%)
Length = 847
Score = 120 bits (300), Expect = 1e-27
Identities = 86/299 (28%), Positives = 135/299 (44%), Gaps = 1/299 (0%)
Frame = +3
Query: 1 MINVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKT 60
++ +L + L A+ + YW S P T++PK + D++ P + KT
Sbjct: 75 IVLALLCVTLKGADASQSAKDYWHSVFPNTRLPKPLWDLVLP--------------SPKT 212
Query: 61 RIVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSD-HGPTFLPG 119
+ PI ++ + + E DLH ++ K +D T
Sbjct: 213 NM-----PI----------KVEEEKQYWTLFFEHDLHPRKIMHLGLHKHNDTKNSTISSW 347
Query: 120 DVANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMID 179
S PF + + +++ + C KGEEK C TSL+SM+
Sbjct: 348 ARTTSQPFGAWLHAKVEERYNLDE-------------VCGKAAAKGEEKFCATSLQSMMG 488
Query: 180 FTTLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDA 239
F KLG N+ +S+ + QYV+ V K+GE V+CH+ N+ VFYCH+ +A
Sbjct: 489 FAISKLGKNIKAISSSFAQDH--DQYVVEE-VNKIGEK-AVMCHRLNFENVVFYCHQINA 656
Query: 240 TKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
T Y VPL +DG++ KA+ ICH DT + ++VLKV+ GT PVCH + + W
Sbjct: 657 TTTYMVPLVASDGTKAKALTICHHDTRGMD-PIVVYEVLKVKTGTVPVCHFVGNKAIAW 830
>TC217024 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzyme 1 beta
subunit precursor , partial (33%)
Length = 958
Score = 119 bits (299), Expect = 1e-27
Identities = 72/215 (33%), Positives = 106/215 (48%), Gaps = 6/215 (2%)
Frame = +1
Query: 90 FLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDVANSIPFSSNKLENILNYFSIKQGSTESE 149
F E L GT + + +FLP + +PFSS K++ + F + + S+ +
Sbjct: 85 FFRETMLKEGTVMPMPDIRDKMPKRSFLPRAILTKLPFSSAKVDELKRVFKVSENSSMDK 264
Query: 150 IVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNVDTVSTEVNGESGLQQYVIAN 209
++ +++ ECE GE K CV S+E MIDF+T LG NV +TE G + V+
Sbjct: 265 MIMDSLGECERAPSVGETKRCVASVEDMIDFSTSVLGRNVAVWTTE--NVKGSNKNVMVG 438
Query: 210 GVKKMGENNL---VVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVK---AVAICHS 263
VK M + V CH+ +PY ++YCH +VY L + S+ K VAICH
Sbjct: 439 RVKGMNGGKVTKSVSCHQSLFPYLLYYCHSLPKVRVYEANLLDPE-SKAKINHGVAICHL 615
Query: 264 DTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
DT+ WS H AF L G VCH + + + W
Sbjct: 616 DTTAWSPTHGAFLALGSGPGRIEVCHWIFENDLTW 720
>TC214787 UP|Q06765 (Q06765) ADR6 protein (Sali5-4a protein), complete
Length = 1150
Score = 117 bits (294), Expect = 5e-27
Identities = 88/297 (29%), Positives = 137/297 (45%)
Frame = +3
Query: 2 INVVLQLALVATHATLPPELYWKSKLPTTQIPKAITDILHPMDDGKISVDGKISVNGKTR 61
++V+ L L A + E +W + P T IP ++ D+L P G SV
Sbjct: 84 LSVLFTLGL-ARESHARDEDFWHAVWPNTPIPSSLRDLLKP---------GPASV----- 218
Query: 62 IVYDAAPIYFYENDANEAELHDNRNLAMFLLEKDLHHGTKFNIQFTKTSDHGPTFLPGDV 121
E D + ++ + + F ++DLH G +QF+K P P V
Sbjct: 219 -----------EIDDHPMQIEETQYPKTFFYKEDLHPGKTMKVQFSKP----PFQQPWGV 353
Query: 122 ANSIPFSSNKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFT 181
+ IK + E + + EA I+GEEK C SL ++I F
Sbjct: 354 GTWLK-------------EIKDTTKEGYSFEELCIKKEA--IEGEEKFCAKSLGTVIGFA 488
Query: 182 TLKLGNNVDTVSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATK 241
KLG N+ +S+ + QY + GV+ +G+ V+CH+ N+ AVFYCH+ T
Sbjct: 489 ISKLGKNIQVLSSSFVNKQ--DQYTV-EGVQNLGDK-AVMCHRLNFRTAVFYCHEVRETT 656
Query: 242 VYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ VPL DG++ +A+AICHS+TS + + Q++ V GT PVCH L ++W
Sbjct: 657 AFMVPLVAGDGTKTQALAICHSNTSGMN-HQMLHQLMGVDPGTNPVCHFLGSKAILW 824
>TC234549 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (14%)
Length = 791
Score = 109 bits (273), Expect = 1e-24
Identities = 57/171 (33%), Positives = 90/171 (52%), Gaps = 8/171 (4%)
Frame = +1
Query: 136 LNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLG----NNVDT 191
L FSI + S + +++T+ +C I GE K+C TSLESM++F +G +N+ T
Sbjct: 25 LQLFSISEDSPXANAMRDTLDQCXXEPITGETKICATSLESMLEFVGKIIGLETKHNIIT 204
Query: 192 VSTEVNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHK-TDATKVYSVPL--- 247
+ LQ++ I + + + V CH YPYA++YCH +KV+ V L
Sbjct: 205 TTLPTASGVPLQKFTILEVSEDINASKWVACHPLPYPYAIYYCHFIATGSKVFKVSLGSE 384
Query: 248 EGADGSRVKAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
D +++A+ ICH DTS WS H+ F+ L ++ G VCH ++W
Sbjct: 385 NNGDDDKIEALGICHLDTSDWSPNHIIFRQLGIKPGKDSVCHFFTIKHLMW 537
>BQ273716 weakly similar to SP|Q08298|RD22_ Dehydration-responsive protein
RD22 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (13%)
Length = 419
Score = 106 bits (265), Expect = 1e-23
Identities = 42/83 (50%), Positives = 58/83 (69%)
Frame = +1
Query: 219 LVVCHKRNYPYAVFYCHKTDATKVYSVPLEGADGSRVKAVAICHSDTSQWSLKHLAFQVL 278
++VCH +YPY VF CH+ T + +PLEG DG+RVKA A+CH DTS+W H+ Q+L
Sbjct: 70 VIVCHPMDYPYVVFMCHEISNTTAHFMPLEGEDGTRVKAAAVCHKDTSEWDPNHVFLQML 249
Query: 279 KVQQGTFPVCHILQQGQVVWFSK 301
K + G PVCHI +G ++WF+K
Sbjct: 250 KTKPGAAPVCHIFPEGHLLWFAK 318
>TC206937 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzyme 1 beta
subunit precursor , partial (23%)
Length = 537
Score = 102 bits (254), Expect = 2e-22
Identities = 58/172 (33%), Positives = 85/172 (48%), Gaps = 3/172 (1%)
Frame = +2
Query: 130 NKLENILNYFSIKQGSTESEIVKNTISECEAYGIKGEEKLCVTSLESMIDFTTLKLGNNV 189
+K++ + F + +++K+++ ECE GE K CV SLE MIDF T LG NV
Sbjct: 5 SKIDELKQVFKASDNGSMEKMMKDSLEECERAPSSGETKRCVGSLEDMIDFATSVLGRNV 184
Query: 190 DTVSTE-VNGESGLQQYVIANGVKKMGENNLVVCHKRNYPYAVFYCHKTDATKVYSVP-L 247
+T+ VNG G+ V CH+ +PY ++YCH +VY L
Sbjct: 185 AVRTTQNVNGSKKSVVVGPVRGINGGKVTQSVSCHQSLFPYLLYYCHAVPKVRVYEADLL 364
Query: 248 EGADGSRV-KAVAICHSDTSQWSLKHLAFQVLKVQQGTFPVCHILQQGQVVW 298
+ +++ + VAICH DTS WS H AF L G VCH + + + W
Sbjct: 365 DPKTKAKINRGVAICHLDTSDWSPTHGAFLSLGSVPGRIEVCHWIFENDMAW 520
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,149,600
Number of Sequences: 63676
Number of extensions: 163245
Number of successful extensions: 961
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 899
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 912
length of query: 301
length of database: 12,639,632
effective HSP length: 97
effective length of query: 204
effective length of database: 6,463,060
effective search space: 1318464240
effective search space used: 1318464240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC127429.4


