

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127429.2 + phase: 2 /pseudo
(500 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
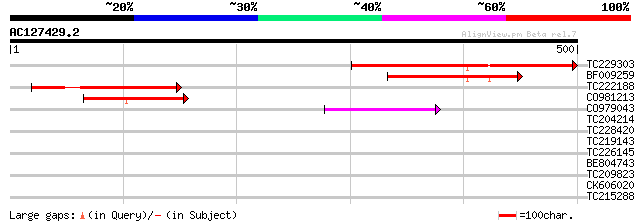
Score E
Sequences producing significant alignments: (bits) Value
TC229303 similar to GB|AAL91627.1|19715609|AY075613 AT3g59630/T1... 332 3e-91
BF009259 199 2e-51
TC222188 similar to GB|AAL91627.1|19715609|AY075613 AT3g59630/T1... 192 2e-49
CO981213 76 3e-14
CO979043 50 3e-06
TC204214 homologue to GB|BAC22311.1|24414062|AP003884 60S riboso... 36 0.046
TC228420 similar to GB|AAR24677.1|38603864|BT010899 At3g62040 {A... 33 0.39
TC219143 similar to UP|Q9FJJ8 (Q9FJJ8) Arabidopsis thaliana geno... 30 2.5
TC226145 weakly similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine p... 30 2.5
BE804743 weakly similar to PIR|T07642|T076 pEARLI 1 protein homo... 29 4.3
TC209823 29 5.6
CK606020 28 7.3
TC215288 UP|Q94C02 (Q94C02) Myo-inositol-1-phosphate synthase ,... 28 9.5
>TC229303 similar to GB|AAL91627.1|19715609|AY075613 AT3g59630/T16L24_180
{Arabidopsis thaliana;} , partial (22%)
Length = 1073
Score = 332 bits (850), Expect = 3e-91
Identities = 166/201 (82%), Positives = 182/201 (89%), Gaps = 2/201 (0%)
Frame = +2
Query: 302 AGYLHIINQMMGLITAAGKKAYTLVMGKPNPAKLANFPECDVFLYVSCAQTALLDSKDYL 361
AGYLHIINQMM LIT AGKKAYTLVMG+PNPAKLANFPECDVFLYVSCAQTALLDSK+YL
Sbjct: 2 AGYLHIINQMMELITGAGKKAYTLVMGRPNPAKLANFPECDVFLYVSCAQTALLDSKEYL 181
Query: 362 APVITPFEATIAFNRGSQWTGAYVMGFGDLINLPQVQVGNQ--EEARFSFLKGGYVEDFE 419
APVITPFEA IAFNRGSQWTGAYVM F DLINLPQ+ VGNQ EEARFSFLKGGYVEDFE
Sbjct: 182 APVITPFEAMIAFNRGSQWTGAYVMEFRDLINLPQMGVGNQEEEEARFSFLKGGYVEDFE 361
Query: 420 NQENNVEEEREDLALANATEKALQLRDNCNSIIKGGAKSGAEFFANRTYQGLNIPDDNTT 479
NQE NVEEERE LAL NATEKALQLR+N N ++KG A+SGAEF ANR+YQGLN+ +NT+
Sbjct: 362 NQE-NVEEERETLALVNATEKALQLRNNSNVLMKGSARSGAEFLANRSYQGLNMSSENTS 538
Query: 480 PQSFVIGRKGRAAGYEDENNK 500
P+ ++IGR+GRA+GYEDE NK
Sbjct: 539 PEPYLIGRRGRASGYEDEKNK 601
>BF009259
Length = 379
Score = 199 bits (507), Expect = 2e-51
Identities = 103/125 (82%), Positives = 110/125 (87%), Gaps = 6/125 (4%)
Frame = +3
Query: 334 KLANFPECDVFLYVSCAQTALLDSKDYLAPVITPFEATIAFNRGSQWTGAYVMGFGDLIN 393
KLANFPECDVFLYVSCAQTALLDSK+YLAPVITPFEA IAFNRGSQWTGAYVM F DLIN
Sbjct: 3 KLANFPECDVFLYVSCAQTALLDSKEYLAPVITPFEAMIAFNRGSQWTGAYVMEFRDLIN 182
Query: 394 LPQVQVGNQ--EEARFSFLKGGYVEDFENQ----ENNVEEEREDLALANATEKALQLRDN 447
LPQ++VGNQ EEARFSFLKGGYVEDFENQ NVEEERE LAL NATEKALQL++N
Sbjct: 183 LPQMEVGNQEEEEARFSFLKGGYVEDFENQACISSENVEEERETLALVNATEKALQLQNN 362
Query: 448 CNSII 452
N ++
Sbjct: 363 SNVLM 377
>TC222188 similar to GB|AAL91627.1|19715609|AY075613 AT3g59630/T16L24_180
{Arabidopsis thaliana;} , partial (18%)
Length = 631
Score = 192 bits (489), Expect = 2e-49
Identities = 95/132 (71%), Positives = 108/132 (80%)
Frame = +1
Query: 20 FFLCEFQFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSGDEPDIGLFVMADTAYGSCCVD 79
F QFPD+LLK STRVV+ LR L+S ++GLFVMADTAYGSCC+D
Sbjct: 271 FIKVALQFPDNLLKHSTRVVTALRNTLQST------------EVGLFVMADTAYGSCCID 414
Query: 80 EVGASHINADCVIHYGHTCFSPTTTLPAFFVFGKASIGLADCVESMSQYALTSSKPIMVL 139
EVGASHINADCVIHYGHTCFSPTTTLPAF VFGKASI +ADCVESMS+YALT+SKPIMVL
Sbjct: 415 EVGASHINADCVIHYGHTCFSPTTTLPAFCVFGKASICIADCVESMSKYALTNSKPIMVL 594
Query: 140 YGLEYAHSIQQV 151
+GLEYAHS+Q++
Sbjct: 595 FGLEYAHSMQRI 630
>CO981213
Length = 710
Score = 76.3 bits (186), Expect = 3e-14
Identities = 36/94 (38%), Positives = 58/94 (61%), Gaps = 2/94 (2%)
Frame = +3
Query: 66 FVMADTAYGSCCVDEVGASHINADCVIHYGHTCFSP--TTTLPAFFVFGKASIGLADCVE 123
FV+AD YG+CCVD++ AS + AD +IHYGH+C P TT+P +VF I + V+
Sbjct: 285 FVLADVTYGACCVDDLAASALGADLLIHYGHSCLVPIDATTIPCLYVFVDIKIDVPHFVD 464
Query: 124 SMSQYALTSSKPIMVLYGLEYAHSIQQVRKALLE 157
++S L +K ++V +++A +I+ + L E
Sbjct: 465 TLSLNLLAQTKTLVVAGTIQFASAIRAAKPQLEE 566
>CO979043
Length = 764
Score = 49.7 bits (117), Expect = 3e-06
Identities = 27/104 (25%), Positives = 58/104 (54%), Gaps = 1/104 (0%)
Frame = -3
Query: 278 RRYYLVERAKDANIVGILVGTLGVAGYLHIINQMMGLITAAGKKAYTLVMGKPNPAKLAN 337
R+ + + ++A G+++GTLG G I+ ++ ++ G ++M + +PA++A
Sbjct: 618 RKNAIFKAREEARSWGLVLGTLGRQGNPRILERLEKMMRDRGFDYTVVLMSEMSPARIAL 439
Query: 338 FPEC-DVFLYVSCAQTALLDSKDYLAPVITPFEATIAFNRGSQW 380
F + D ++ ++C + ++ + ++ PV+TPFEA IA W
Sbjct: 438 FEDSIDAWIQIACPRLSIDWGEAFVKPVLTPFEADIALGVIPGW 307
>TC204214 homologue to GB|BAC22311.1|24414062|AP003884 60S ribosomal protein
L7A {Oryza sativa (japonica cultivar-group);} , partial
(7%)
Length = 697
Score = 35.8 bits (81), Expect = 0.046
Identities = 16/19 (84%), Positives = 16/19 (84%)
Frame = +2
Query: 376 RGSQWTGAYVMGFGDLINL 394
RGSQWTGAYVM F D INL
Sbjct: 554 RGSQWTGAYVMEFQDPINL 610
>TC228420 similar to GB|AAR24677.1|38603864|BT010899 At3g62040 {Arabidopsis
thaliana;} , partial (55%)
Length = 810
Score = 32.7 bits (73), Expect = 0.39
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 5/81 (6%)
Frame = +2
Query: 122 VESMSQYALTSSKPIMVLYGLEYAHSIQQVRKALLESS-----TFCTSDPKSEVHFADVP 176
V S+ QY+LT + + ++AH+++ + + LE F T +P +++ DVP
Sbjct: 125 VRSIPQYSLT*KCYLQIFTNADHAHAVKVLNRLGLEDCFEGIICFETLNPPKQINCMDVP 304
Query: 177 SPYMFPSKDTKKLNDLQEETC 197
+ D L DL E C
Sbjct: 305 N-------DNHVLTDLTENGC 346
>TC219143 similar to UP|Q9FJJ8 (Q9FJJ8) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K19B1, partial (88%)
Length = 689
Score = 30.0 bits (66), Expect = 2.5
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 6/86 (6%)
Frame = +1
Query: 20 FFLCEFQFPDDLLKDSTRVVSVLRKRLRSLRESDTTGSG-DEPDIGLFVMA-----DTAY 73
FF C + +L++ VV V+ + E G G DE ++ + VM D Y
Sbjct: 394 FFPCRKRCI*NLMRTRHWVVVVVEEESIMEEERVAEGGGVDEEEVDVVVMGEEESGDIKY 573
Query: 74 GSCCVDEVGASHINADCVIHYGHTCF 99
SCC E + N C++ +CF
Sbjct: 574 NSCCWSEFYSDI*NVFCILVLLISCF 651
>TC226145 weakly similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein
kinase-like protein (CBL-interacting protein kinase 5),
partial (15%)
Length = 890
Score = 30.0 bits (66), Expect = 2.5
Identities = 19/42 (45%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Frame = +1
Query: 390 DLINLPQVQVGNQEEARFSFLKGGYVED---FENQENNVEEE 428
D++ P QVG FS +K YVED F++ ENN EEE
Sbjct: 31 DIMRDPWFQVGFMRPIAFS-IKESYVEDNIDFDDVENNQEEE 153
>BE804743 weakly similar to PIR|T07642|T076 pEARLI 1 protein homolog T1P17.90
- Arabidopsis thaliana, partial (41%)
Length = 349
Score = 29.3 bits (64), Expect = 4.3
Identities = 21/76 (27%), Positives = 37/76 (48%), Gaps = 5/76 (6%)
Frame = +1
Query: 67 VMADTAYGSCCVDEVGASHINADCVIHYGH-TCFSPTTTL--PAFFVFGKASIG-LADCV 122
V+ D G CC+ +H++ IH+G + +PTT + + SIG + CV
Sbjct: 25 VV*DGFQGKCCIHLCCTTHVSLFAFIHHGELSNLNPTTNAFSKRYMSYKCTSIGCVCQCV 204
Query: 123 ESMSQYA-LTSSKPIM 137
E Y +T++ P++
Sbjct: 205 EPGQCYTRITTNPPLL 252
>TC209823
Length = 1092
Score = 28.9 bits (63), Expect = 5.6
Identities = 22/73 (30%), Positives = 31/73 (42%)
Frame = +1
Query: 221 KLLEGQSMEDYSLFWIGHDNSAFANVVLRFNACEIVRYDATKSQMVADLSQQRRILKRRY 280
K L+G S+ S I H NS FA R N CE V + ++ A LS + L+
Sbjct: 445 KNLKGDSVNKESQRRISHHNSDFAVSTSRLNDCEAVVHISSYEAHKAPLSDILQCLENNG 624
Query: 281 YLVERAKDANIVG 293
+ A + G
Sbjct: 625 LYLLNASSSETFG 663
>CK606020
Length = 340
Score = 28.5 bits (62), Expect = 7.3
Identities = 14/54 (25%), Positives = 25/54 (45%)
Frame = -3
Query: 183 SKDTKKLNDLQEETCGCGDNSSSDGASGAAYSIGGLTWKLLEGQSMEDYSLFWI 236
S + + + + +SSS +S ++ S G ++ L G+S D L WI
Sbjct: 287 SSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSDGSISSSLATGESGSDIGLNWI 126
>TC215288 UP|Q94C02 (Q94C02) Myo-inositol-1-phosphate synthase , complete
Length = 1939
Score = 28.1 bits (61), Expect = 9.5
Identities = 17/51 (33%), Positives = 23/51 (44%), Gaps = 2/51 (3%)
Frame = -2
Query: 130 LTSSKPIMVLYGLEYAHSIQQVRKALLESST--FCTSDPKSEVHFADVPSP 178
L S I V+ E+ H + LL+ C S+VH AD+PSP
Sbjct: 786 LVGSNEIRVIDSGEWKHGFHVRPQLLLQIDVKHLCPGHGISQVHVADIPSP 634
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,078,544
Number of Sequences: 63676
Number of extensions: 278721
Number of successful extensions: 1193
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 1178
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1186
length of query: 500
length of database: 12,639,632
effective HSP length: 101
effective length of query: 399
effective length of database: 6,208,356
effective search space: 2477134044
effective search space used: 2477134044
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC127429.2


