

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.2 - phase: 0
(270 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
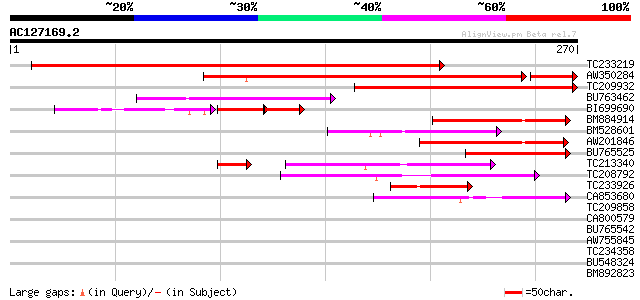
Score E
Sequences producing significant alignments: (bits) Value
TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance... 293 7e-80
AW350284 similar to GP|22946224|gb| CG31868-PA {Drosophila melan... 222 2e-62
TC209932 162 2e-40
BU763462 weakly similar to GP|9757923|dbj| laccase (diphenol oxi... 71 5e-13
BI699690 49 2e-11
BM884914 59 2e-09
BM528601 57 9e-09
AW201846 57 9e-09
BU765525 55 3e-08
TC213340 50 7e-08
TC208792 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp prote... 53 1e-07
TC233926 48 4e-06
CA853680 42 4e-04
TC209858 38 0.006
CA800579 29 0.008
BU765542 37 0.012
AW755845 37 0.012
TC234358 36 0.021
BU548324 35 0.047
BM892823 similar to GP|22946353|gb CG6214-PA {Drosophila melanog... 34 0.061
>TC233219 weakly similar to UP|Q8LKI9 (Q8LKI9) NBS/LRR resistance
protein-like protein (Fragment), partial (74%)
Length = 781
Score = 293 bits (749), Expect = 7e-80
Identities = 139/197 (70%), Positives = 164/197 (82%)
Frame = +3
Query: 11 VTEHIQELAPRCMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFS 70
+TE IQE+ RCMLFADD+VL+GESRE++N RLETWR+ALE +GFRLSRSK+EY+EC F+
Sbjct: 189 LTEQIQEIVSRCMLFADDIVLLGESREKLNERLETWRRALETHGFRLSRSKSEYMECKFN 368
Query: 71 GRRSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLKWRRDSDVLCD 130
RR S EVK+ DHIIPQVTR KYLGS +Q+DGEIE DV+HRIQA W+KWR+ S VLCD
Sbjct: 369 KRRRVSNSEVKIGDHIIPQVTRFKYLGSVIQDDGEIEGDVNHRIQA*WMKWRKASGVLCD 548
Query: 131 KKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRND 190
KVP+KLKGKFYR A RPA++YG ECWAVKSQHEN+V VA RMLRWM KTR D+ RN+
Sbjct: 549 AKVPIKLKGKFYRTAVRPAILYGTECWAVKSQHENKVGVAXXRMLRWMCGKTRQDKIRNE 728
Query: 191 TIRKRVEVAPIVEKLVE 207
+RV VAPIVEK+VE
Sbjct: 729 AXXERVGVAPIVEKMVE 779
>AW350284 similar to GP|22946224|gb| CG31868-PA {Drosophila melanogaster},
partial (1%)
Length = 767
Score = 222 bits (566), Expect(2) = 2e-62
Identities = 109/160 (68%), Positives = 124/160 (77%), Gaps = 6/160 (3%)
Frame = -1
Query: 93 SKYLGSFVQNDGEIEADVS------HRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRPAF 146
SK F N +E +S HRIQAGW+KWR+ S VLCD KVP+KLKGKFYR A
Sbjct: 665 SKKNKCFFNNTEVLEVFISV*LFSFHRIQAGWMKWRKTSGVLCDAKVPIKLKGKFYRTAV 486
Query: 147 RPALMYGRECWAVKSQHENQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRVEVAPIVEKLV 206
RPA++YG ECWAVKSQHEN+V VAEMRMLRWM KTR D+ RN+ IR+RV VAPIVEK+V
Sbjct: 485 RPAILYGTECWAVKSQHENKVGVAEMRMLRWMCGKTRQDKIRNEAIRERVGVAPIVEKMV 306
Query: 207 ENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKT 246
ENRLRWFGHVERRPVD+VVRR+ QME Q GRGRP+KT
Sbjct: 305 ENRLRWFGHVERRPVDSVVRRIDQMERRQTIGGRGRPKKT 186
Score = 34.3 bits (77), Expect(2) = 2e-62
Identities = 15/22 (68%), Positives = 18/22 (81%)
Frame = -2
Query: 249 ETIRKDLEVNELDPNLVYDRTL 270
E I+KDLE+N LD N+V DRTL
Sbjct: 184 EVIKKDLEINGLDRNIVLDRTL 119
>TC209932
Length = 829
Score = 162 bits (409), Expect = 2e-40
Identities = 79/106 (74%), Positives = 91/106 (85%)
Frame = +2
Query: 165 NQVSVAEMRMLRWMSSKTRHDRTRNDTIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAV 224
N+V VAEMRMLRWM KTR D+ RN+ IR+RV VAPIVEK+VENRLRWFGHVERRPVD+V
Sbjct: 224 NKVGVAEMRMLRWMCGKTRQDKIRNEAIRERVGVAPIVEKMVENRLRWFGHVERRPVDSV 403
Query: 225 VRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYDRTL 270
VRRV QME Q RGRGRP+KTIRE I+KDLE+N+LD ++V DRTL
Sbjct: 404 VRRVDQMERRQTIRGRGRPKKTIREVIKKDLELNDLDRSMVLDRTL 541
>BU763462 weakly similar to GP|9757923|dbj| laccase (diphenol oxidase)
{Arabidopsis thaliana}, partial (4%)
Length = 421
Score = 71.2 bits (173), Expect = 5e-13
Identities = 38/95 (40%), Positives = 55/95 (57%)
Frame = +1
Query: 61 KTEYIECNFSGRRSRSTLEVKVEDHIIPQVTRSKYLGSFVQNDGEIEADVSHRIQAGWLK 120
KT+Y+ NFS + + LEVK+ D IP V++ KY +QN I D +H++Q G LK
Sbjct: 1 KTKYMHSNFSKK*EENELEVKMGD-AIP*VSKFKYFEPILQNSW*INEDGTHKMQLGCLK 177
Query: 121 WRRDSDVLCDKKVPLKLKGKFYRPAFRPALMYGRE 155
+ S + CD KVP +K +FY P ++YG E
Sbjct: 178 EAKASRINCDHKVPTNIKAQFYCTVIHPNILYGNE 282
>BI699690
Length = 407
Score = 49.3 bits (116), Expect(3) = 2e-11
Identities = 35/83 (42%), Positives = 48/83 (57%), Gaps = 6/83 (7%)
Frame = -1
Query: 22 CMLFADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVK 81
CML ADD+ + ES G + W+Q+ F LSRSKT+Y+ C+FS ++ E K
Sbjct: 347 CMLSADDIFFIEESHNMRFGH-KLWKQS-----FHLSRSKTKYMYCSFSKKQE----EYK 198
Query: 82 VED--HIIPQVT----RSKYLGS 98
ED +IPQV+ + KYLGS
Sbjct: 197 *EDGEDVIPQVSKFKFKFKYLGS 129
Score = 30.4 bits (67), Expect(3) = 2e-11
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = -3
Query: 122 RRDSDVLCDKKVPLKLKGK 140
+R V+CD KVP+KLKGK
Sbjct: 57 KRQLGVICDHKVPIKLKGK 1
Score = 25.0 bits (53), Expect(3) = 2e-11
Identities = 11/24 (45%), Positives = 17/24 (70%)
Frame = -2
Query: 100 VQNDGEIEADVSHRIQAGWLKWRR 123
++N G++ DVSH IQA LK ++
Sbjct: 124 MKNYGKMNEDVSHEIQAKRLKCKK 53
>BM884914
Length = 423
Score = 58.9 bits (141), Expect = 2e-09
Identities = 30/66 (45%), Positives = 45/66 (67%)
Frame = +1
Query: 202 VEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVNELD 261
VE ++EN+L+W+ +RP++ +VR+VY S KR RGRPR T+ E I+KDL VN +
Sbjct: 7 VENVIENQLKWWFEYVQRPLETLVRKVYHTVFSLSKRCRGRPR-TLMEIIKKDLMVNNIP 183
Query: 262 PNLVYD 267
LV++
Sbjct: 184 EILVFN 201
>BM528601
Length = 426
Score = 57.0 bits (136), Expect = 9e-09
Identities = 38/88 (43%), Positives = 52/88 (58%), Gaps = 5/88 (5%)
Frame = +3
Query: 152 YGRECWAVKSQHENQVSVA---EMRML--RWMSSKTRHDRTRNDTIRKRVEVAPIVEKLV 206
Y +C AVK E++ EMRML MS R ++ N+ IR++V VA I EK+V
Sbjct: 156 YCTKCSAVKCHQEHKTKCC*ELEMRML**LMMSIHARIEKM-NECIREKVGVALIQEKMV 332
Query: 207 ENRLRWFGHVERRPVDAVVRRVYQMEDS 234
E LRWF HV RRP++A + +V +E S
Sbjct: 333 EMHLRWFDHVPRRPIEAQMSKVD*IEGS 416
>AW201846
Length = 416
Score = 57.0 bits (136), Expect = 9e-09
Identities = 29/71 (40%), Positives = 48/71 (66%)
Frame = +2
Query: 196 VEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDL 255
++V PI EK+ E + WFG ++ +P++ +VRR+ + S VK+ RG PR T +E I++DL
Sbjct: 50 IDVMPIKEKMTEK*VGWFGCMQ*KPLEVLVRRMDCIIFSLVKKSRGEPR-T*KEIIKRDL 226
Query: 256 EVNELDPNLVY 266
VN++ N V+
Sbjct: 227 MVNDVPENFVF 259
>BU765525
Length = 421
Score = 55.1 bits (131), Expect = 3e-08
Identities = 24/50 (48%), Positives = 36/50 (72%)
Frame = +3
Query: 218 RRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYD 267
RRP++A +R + M S VKRGRGRPR+T+ E ++KD VN + +LV++
Sbjct: 18 RRPMEAPMRNINYMVFSPVKRGRGRPRRTLEEVVKKDFMVNNISDDLVFN 167
>TC213340
Length = 501
Score = 50.4 bits (119), Expect(2) = 7e-08
Identities = 32/102 (31%), Positives = 49/102 (47%), Gaps = 2/102 (1%)
Frame = -1
Query: 132 KVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVS--VAEMRMLRWMSSKTRHDRTRN 189
KV LK +FY P ++Y ECW K HE +VS ++ W K++ +
Sbjct: 381 KVLTNLKREFYCTVIEPTILYSSECWGSKG*HEKKVSNRNENTKIDEWPHKKSQDTK--- 211
Query: 190 DTIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQM 231
+ + V I E + N + F HV++RP++A V RV M
Sbjct: 210 *LYKGNISVTTIEEWTIGN*IICFRHVQKRPLEAPVGRVNLM 85
Score = 23.1 bits (48), Expect(2) = 7e-08
Identities = 9/16 (56%), Positives = 13/16 (81%)
Frame = -3
Query: 100 VQNDGEIEADVSHRIQ 115
VQN+ EI DV+H++Q
Sbjct: 478 VQNNVEINKDVTHKMQ 431
>TC208792 homologue to UP|Q9S834 (Q9S834) ATP-dependent Clp protease subunit
ClpP (NClpP1) (ATP-dependent Clp protease proteolytic
subunit ClpP5) , partial (14%)
Length = 739
Score = 53.1 bits (126), Expect = 1e-07
Identities = 40/124 (32%), Positives = 61/124 (48%), Gaps = 1/124 (0%)
Frame = -2
Query: 130 DKKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVSVAEMR-MLRWMSSKTRHDRTR 188
+KKVPLKLKGK + R ++YG + W VK Q + +VAE R M + T
Sbjct: 597 NKKVPLKLKGKLQDTSIRSMMVYGTKEWVVKGQQ*RKHNVAETR*D*E*MQQRESWSNT- 421
Query: 189 NDTIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIR 248
++V+ R FG V ++ +VRRV +E+S + RGRG+ TI
Sbjct: 420 ------------YCGEVVKYHHR*FGSVWISRIETLVRRVD*IEESPITRGRGKL*NTIE 277
Query: 249 ETIR 252
+ ++
Sbjct: 276 KLLK 265
>TC233926
Length = 570
Score = 48.1 bits (113), Expect = 4e-06
Identities = 21/39 (53%), Positives = 29/39 (73%)
Frame = +3
Query: 182 TRHDRTRNDTIRKRVEVAPIVEKLVENRLRWFGHVERRP 220
TR DR +ND IR + V PI +K+ +N LRWFGH++R+P
Sbjct: 48 TRKDRVQND*IRD-ISVVPIKDKMTQNGLRWFGHMQRKP 161
>CA853680
Length = 575
Score = 41.6 bits (96), Expect = 4e-04
Identities = 28/96 (29%), Positives = 55/96 (57%), Gaps = 2/96 (2%)
Frame = +1
Query: 174 MLRWMSSKTRHDRTRNDTIRKRVEVAPIVEKLVENRLRWF--GHVERRPVDAVVRRVYQM 231
ML M KTR D+ R+D I ++VA EK+ +N + G+++ + ++ +V R
Sbjct: 70 MLG*MCGKTRKDKIRSDRI**NIDVALDEEKMTKNS*NSYICGNLKHQ-LNYIVNR---- 234
Query: 232 EDSQVKRGRGRPRKTIRETIRKDLEVNELDPNLVYD 267
+K+ +G+PR+T+ + + +DL + + NLV++
Sbjct: 235 ---PMKKDQGKPRRTLEKIVTRDLMMASIFENLVFN 333
>TC209858
Length = 1340
Score = 37.7 bits (86), Expect = 0.006
Identities = 19/34 (55%), Positives = 23/34 (66%)
Frame = -3
Query: 128 LCDKKVPLKLKGKFYRPAFRPALMYGRECWAVKS 161
LC + VPLK KGKFY P R A+ G +C A+KS
Sbjct: 285 LC-QNVPLKTKGKFYHPNIRHAM*CGTKCRAIKS 187
Score = 34.7 bits (78), Expect = 0.047
Identities = 33/118 (27%), Positives = 47/118 (38%), Gaps = 2/118 (1%)
Frame = -2
Query: 109 DVSHRIQAGWLKWRRDSDVLCDKKVPLKLKGKFYRPAFRPALMYGRECWAVKSQHENQVS 168
DV+HRI AG +KW+ C K+ Y+ W H+
Sbjct: 334 DVNHRIHAGRMKWKILFMPECTT*NQRKILPSKYKTC--------NVVWN*MPCHKKLTK 179
Query: 169 VAEMRMLR--WMSSKTRHDRTRNDTIRKRVEVAPIVEKLVENRLRWFGHVERRPVDAV 224
R + T DR RN I ++V +AP+VEK++ RWF R VD +
Sbjct: 178 RINSM**R*DYYIGCTGQDRIRNVRIGEKVGIAPMVEKML*YYYRWFLENPVRRVDQI 5
>CA800579
Length = 409
Score = 28.9 bits (63), Expect(2) = 0.008
Identities = 14/22 (63%), Positives = 16/22 (72%)
Frame = +3
Query: 226 RRVYQMEDSQVKRGRGRPRKTI 247
RRV + S VKRGRGR RKT+
Sbjct: 342 RRVIPIVFSTVKRGRGRSRKTL 407
Score = 27.3 bits (59), Expect(2) = 0.008
Identities = 11/20 (55%), Positives = 17/20 (85%)
Frame = +2
Query: 208 NRLRWFGHVERRPVDAVVRR 227
NRLR FG+++R P+ A+V+R
Sbjct: 281 NRLR*FGYMQRGPLKALVKR 340
>BU765542
Length = 427
Score = 36.6 bits (83), Expect = 0.012
Identities = 17/33 (51%), Positives = 22/33 (66%)
Frame = +2
Query: 238 RGRGRPRKTIRETIRKDLEVNELDPNLVYDRTL 270
RG+GR RK E+I D EVN L +++Y RTL
Sbjct: 2 RGKGRTRKN*GESITNDSEVNNLSIDMIYSRTL 100
>AW755845
Length = 152
Score = 36.6 bits (83), Expect = 0.012
Identities = 18/44 (40%), Positives = 26/44 (58%), Gaps = 2/44 (4%)
Frame = -2
Query: 78 LEVKVEDHIIP--QVTRSKYLGSFVQNDGEIEADVSHRIQAGWL 119
LE K+ H VT +YLGS +Q GE + DV+ +I +GW+
Sbjct: 136 LEEKIRYHPYQ*YNVT*FRYLGSIIQRKGETKGDVNQKIHSGWM 5
>TC234358
Length = 432
Score = 35.8 bits (81), Expect = 0.021
Identities = 25/68 (36%), Positives = 37/68 (53%)
Frame = -2
Query: 25 FADDVVLVGESREEVNGRLETWRQALEAYGFRLSRSKTEYIECNFSGRRSRSTLEVKVED 84
F DDVVL+ +SRE V + + W + + L RSKT YI + L+VK+
Sbjct: 317 FLDDVVLIEDSREVVYFKHKLWIYNFDTRSYCLRRSKTVYI---LQFQ*EEYKLKVKIR- 150
Query: 85 HIIPQVTR 92
+I+ QV+R
Sbjct: 149 NILSQVSR 126
>BU548324
Length = 695
Score = 34.7 bits (78), Expect = 0.047
Identities = 21/53 (39%), Positives = 31/53 (57%)
Frame = -1
Query: 212 WFGHVERRPVDAVVRRVYQMEDSQVKRGRGRPRKTIRETIRKDLEVNELDPNL 264
WF V +RPV+++V +V QME + + K I + + KDLEVN L N+
Sbjct: 395 WFESV*KRPVESLVTKVDQMEGNPIVLEVEESEKKIGK-LNKDLEVNGLYINM 240
>BM892823 similar to GP|22946353|gb CG6214-PA {Drosophila melanogaster},
partial (0%)
Length = 421
Score = 34.3 bits (77), Expect = 0.061
Identities = 16/28 (57%), Positives = 21/28 (74%)
Frame = +1
Query: 201 IVEKLVENRLRWFGHVERRPVDAVVRRV 228
I EK +E +LR FGHV+R P++A RRV
Sbjct: 70 IEEKKIETQLR*FGHVQRSPLEASRRRV 153
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,315,160
Number of Sequences: 63676
Number of extensions: 141773
Number of successful extensions: 723
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 720
length of query: 270
length of database: 12,639,632
effective HSP length: 96
effective length of query: 174
effective length of database: 6,526,736
effective search space: 1135652064
effective search space used: 1135652064
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC127169.2


