

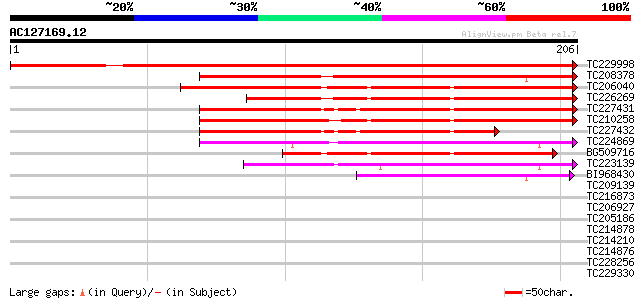
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127169.12 + phase: 0
(206 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229998 weakly similar to PIR|T04232|T04232 pathogenesis-relate... 359 e-100
TC208378 similar to UP|PR1_MEDTR (Q40374) Pathogenesis-related p... 174 3e-44
TC206040 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (... 136 6e-33
TC226269 homologue to UP|Q9XFB4 (Q9XFB4) PR1a precursor, complete 134 4e-32
TC227431 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (... 125 1e-29
TC210258 weakly similar to UP|Q9LPM7 (Q9LPM7) F2J10.6 protein, p... 123 5e-29
TC227432 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (... 91 4e-19
TC224869 weakly similar to UP|ST14_SOLTU (Q41495) STS14 protein ... 86 9e-18
BG509716 similar to GP|4928711|gb|A PR1a precursor {Glycine max}... 82 2e-16
TC223139 weakly similar to UP|Q8LDA3 (Q8LDA3) Sts14, partial (59%) 78 3e-15
BI968430 48 4e-06
TC209139 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein... 31 0.35
TC216873 similar to UP|Q9SCQ0 (Q9SCQ0) RNA helicase-like protein... 30 1.0
TC206927 homologue to UP|Q9ZPK4 (Q9ZPK4) Glutamyl-tRNA reductase... 29 1.3
TC205186 similar to UP|Q8H9B8 (Q8H9B8) Low temperature and salt ... 29 1.7
TC214878 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit bindi... 28 2.2
TC214210 28 2.2
TC214876 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit bindi... 28 2.2
TC228256 weakly similar to UP|Q9ATY4 (Q9ATY4) MAP kinase phospha... 28 2.2
TC229330 similar to GB|AAQ56838.1|34098911|BT010395 At1g16900 {A... 28 2.2
>TC229998 weakly similar to PIR|T04232|T04232 pathogenesis-related protein
homolog F14M19.60 - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (78%)
Length = 915
Score = 359 bits (922), Expect = e-100
Identities = 162/206 (78%), Positives = 179/206 (86%)
Frame = +1
Query: 1 MKSHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQL 60
MK+HL+L I VTFT+ + S P S+SS P L+QQK+PDNE+IY+VSKQL
Sbjct: 7 MKAHLVLFLLMILVTFTSNVNTLSINPKSNSSIPQ------LTQQKRPDNETIYRVSKQL 168
Query: 61 CWNCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFK 120
CW C+ ESLEFLFRHNLVRA+KWELPLMWD+QLEQYARWWA +RK DCK+EHSFPEDGFK
Sbjct: 169 CWGCIAESLEFLFRHNLVRAAKWELPLMWDFQLEQYARWWAGERKADCKLEHSFPEDGFK 348
Query: 121 LGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCAR 180
LGENIYWGSGS WTP+DAV+AWADEEKYYTY TN+CV GQMCGHYTQIVWKSTRRIGCAR
Sbjct: 349 LGENIYWGSGSAWTPSDAVRAWADEEKYYTYATNTCVPGQMCGHYTQIVWKSTRRIGCAR 528
Query: 181 VVCDDGDVFMTCNYDPVGNYVGERPY 206
VVCDDGDVFMTCNYDPVGNYVGERPY
Sbjct: 529 VVCDDGDVFMTCNYDPVGNYVGERPY 606
>TC208378 similar to UP|PR1_MEDTR (Q40374) Pathogenesis-related protein PR-1
precursor, partial (84%)
Length = 779
Score = 174 bits (441), Expect = 3e-44
Identities = 79/138 (57%), Positives = 95/138 (68%), Gaps = 1/138 (0%)
Frame = +1
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+FL N RA PL+WD +L YA+W+A+QR+ DC +EHS GENI+WGS
Sbjct: 160 QFLIPQNAARAVLRLRPLVWDSKLAHYAQWYANQRRNDCALEHS----NGPYGENIFWGS 327
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG-DV 188
G+ W P AV AW +E ++Y Y NSC +GQMCGHYTQIVW +TR+IGCA VVC G
Sbjct: 328 GTGWKPAQAVSAWVEERQWYNYWHNSCANGQMCGHYTQIVWSTTRKIGCASVVCSGGKGT 507
Query: 189 FMTCNYDPVGNYVGERPY 206
FMTCNYDP GNY GERPY
Sbjct: 508 FMTCNYDPPGNYYGERPY 561
>TC206040 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (90%)
Length = 772
Score = 136 bits (343), Expect = 6e-33
Identities = 66/144 (45%), Positives = 92/144 (63%)
Frame = +3
Query: 63 NCMQESLEFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLG 122
N +++ HN R+ L WD + +A+ +A+QRK DC++ HS G + G
Sbjct: 159 NAQDSPADYVNAHNAARSEVGVQNLAWDDTVAAFAQNYANQRKGDCQLIHS--GGGGQYG 332
Query: 123 ENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVV 182
EN+ +G D + TDAVK W DE+ Y Y +NSCV G+ C HYTQ+VW+ + R+GCA+V
Sbjct: 333 ENLAMSTG-DLSGTDAVKLWVDEKSNYDYNSNSCVGGE-CLHYTQVVWRDSVRLGCAKVA 506
Query: 183 CDDGDVFMTCNYDPVGNYVGERPY 206
CD+G F+TCNY P GNYVG+RPY
Sbjct: 507 CDNGGTFITCNYAPPGNYVGQRPY 578
>TC226269 homologue to UP|Q9XFB4 (Q9XFB4) PR1a precursor, complete
Length = 771
Score = 134 bits (336), Expect = 4e-32
Identities = 64/120 (53%), Positives = 84/120 (69%)
Frame = +3
Query: 87 LMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGSGSDWTPTDAVKAWADEE 146
L WD + YA +A+QRK DC++ HS G + GENI +G + + TDAVK W DE+
Sbjct: 255 LAWDDTVAAYAESYANQRKGDCQLIHS----GGEYGENIAMSTG-ELSGTDAVKMWVDEK 419
Query: 147 KYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVFMTCNYDPVGNYVGERPY 206
Y Y +NSCV G+ C HYTQ+VW ++ R+GCA+V CD+G F+TCNYDP GN+VGERPY
Sbjct: 420 SNYDYDSNSCVGGE-CLHYTQVVWANSVRLGCAKVTCDNGGTFITCNYDPPGNFVGERPY 596
>TC227431 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (90%)
Length = 707
Score = 125 bits (314), Expect = 1e-29
Identities = 61/137 (44%), Positives = 92/137 (66%)
Frame = +2
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+++ HN R+ ++WD + +A+ +A+QRK DCK+ HS DG K GEN+ GS
Sbjct: 152 DYVNAHNAARSQVGVPNIVWDNAVAAFAQNYANQRKGDCKLVHS-GGDG-KYGENLA-GS 322
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVF 189
+ + DAV+ W +E+ Y Y +NSCV G+ C HYTQ+VW+++ R+GCA+V C++G F
Sbjct: 323 TGNLSGKDAVQLWVNEKSKYNYNSNSCVGGE-CLHYTQVVWRNSLRLGCAKVRCNNGGTF 499
Query: 190 MTCNYDPVGNYVGERPY 206
+ CNY P GNY+G+RPY
Sbjct: 500 IGCNYAPPGNYIGQRPY 550
>TC210258 weakly similar to UP|Q9LPM7 (Q9LPM7) F2J10.6 protein, partial (66%)
Length = 608
Score = 123 bits (309), Expect = 5e-29
Identities = 62/137 (45%), Positives = 86/137 (62%)
Frame = +1
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+FL HN RA PL W++ L+ YA+ +A++R PDC +EHS GEN+ G
Sbjct: 142 DFLDVHNQARAEVGVGPLSWNHTLQAYAQRYANERIPDCNLEHSMGP----FGENLAEGY 309
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVF 189
G + +DAVK W E+ YY + +N+CV + C HYTQIVW+ + +GCAR C++G VF
Sbjct: 310 G-EMKGSDAVKFWLTEKPYYDHYSNACVHDE-CLHYTQIVWRDSVHLGCARAKCNNGWVF 483
Query: 190 MTCNYDPVGNYVGERPY 206
+ C+Y P GN GERPY
Sbjct: 484 VICSYSPPGNIEGERPY 534
>TC227432 similar to UP|Q9XFB4 (Q9XFB4) PR1a precursor, partial (75%)
Length = 448
Score = 90.9 bits (224), Expect = 4e-19
Identities = 48/109 (44%), Positives = 71/109 (65%)
Frame = +2
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGS 129
+F+ HN R+ ++WD + +A+ +A+QRK DCK+ HS DG K GEN+ GS
Sbjct: 131 DFVNAHNAARSQVGVPNIVWDDTVAAFAQNYANQRKGDCKLVHS-GGDG-KYGENLA-GS 301
Query: 130 GSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGC 178
+ + T+AVK W DE+ Y Y +N+CV G+ C HYTQ+VWK++ R+GC
Sbjct: 302 TGNLSGTNAVKLWVDEKSKYDYNSNTCVGGE-CRHYTQVVWKNSVRLGC 445
>TC224869 weakly similar to UP|ST14_SOLTU (Q41495) STS14 protein precursor,
partial (65%)
Length = 1276
Score = 86.3 bits (212), Expect = 9e-18
Identities = 47/140 (33%), Positives = 67/140 (47%), Gaps = 3/140 (2%)
Frame = +3
Query: 70 EFLFRHNLVRASKWELPLMWDYQLEQYARWWA--SQRKPDCKVEHSFPEDGFKLGENIYW 127
EFL HN RA L W +L + + K C+ + + G N W
Sbjct: 678 EFLEAHNQARAEVGVEALSWSEKLGNVSSLMVRYQRNKKGCEFANLTAS---RYGGNQLW 848
Query: 128 GSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGD 187
++ P V+ W E+K+Y N+CV CG YTQ+VW+++ +GCA+ VC
Sbjct: 849 AGVTEVAPRVVVEEWVKEKKFYVRENNTCVGKHECGVYTQVVWRNSTEVGCAQAVCVKEQ 1028
Query: 188 VFMT-CNYDPVGNYVGERPY 206
+T C YDP GN +GE PY
Sbjct: 1029ASLTICFYDPPGNVIGEIPY 1088
>BG509716 similar to GP|4928711|gb|A PR1a precursor {Glycine max}, partial
(54%)
Length = 294
Score = 82.0 bits (201), Expect = 2e-16
Identities = 44/100 (44%), Positives = 63/100 (63%)
Frame = +3
Query: 100 WASQRKPDCKVEHSFPEDGFKLGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSG 159
+A+QRK DC++ HS G + GEN+ +G D + TDAV+ DE+ Y Y +NSCV G
Sbjct: 6 YANQRKGDCQLIHS--GGGGQYGENLAMSTG-DLSGTDAVRGGGDEKSNYDYNSNSCVGG 176
Query: 160 QMCGHYTQIVWKSTRRIGCARVVCDDGDVFMTCNYDPVGN 199
+ C HYTQ+VW+ + R G A+ ++G +T NY P GN
Sbjct: 177 E-CLHYTQVVWRDSVRXGXAKXAWENGGPXITGNYAPPGN 293
>TC223139 weakly similar to UP|Q8LDA3 (Q8LDA3) Sts14, partial (59%)
Length = 634
Score = 77.8 bits (190), Expect = 3e-15
Identities = 43/123 (34%), Positives = 61/123 (48%), Gaps = 2/123 (1%)
Frame = +2
Query: 86 PLMWDYQLEQYARWWASQRKPDCKVEHSFPEDGFKLGENIYWGSGSDW-TPTDAVKAWAD 144
PL W Q+ A ++ + + G K G N GS TP AV+ W
Sbjct: 5 PLRWSEQVANVTSKLARYQRVKTGCQFANLTAG-KYGANQLLARGSAAVTPRMAVEEWVK 181
Query: 145 EEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDGDVFMT-CNYDPVGNYVGE 203
++++Y + NSC CG YTQ+VW+ + +GCA+ C +T C Y+P GNYVGE
Sbjct: 182 QKQFYFHADNSCAPNHRCGVYTQVVWRKSVELGCAQATCVKEQASLTICFYNPPGNYVGE 361
Query: 204 RPY 206
PY
Sbjct: 362 SPY 370
>BI968430
Length = 401
Score = 47.8 bits (112), Expect = 4e-06
Identities = 26/80 (32%), Positives = 39/80 (48%), Gaps = 1/80 (1%)
Frame = -2
Query: 127 WGSGSDWTPTDAVKAWADEEKYYTYVTNSCVSGQMCGHYTQIVWKSTRRIGCARVVCDDG 186
WGSGS W + A E + + Y ++SC +GQMCG + + GC + C G
Sbjct: 376 WGSGSGWHS*QSDSA*GIEHQRF*YRSSSCHNGQMCGQNCPFGSRDSXTTGCTSIGCTGG 197
Query: 187 -DVFMTCNYDPVGNYVGERP 205
D F + + D G+ G+ P
Sbjct: 196 KDTFRSSS*DRNGSCYGQSP 137
>TC209139 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (45%)
Length = 733
Score = 31.2 bits (69), Expect = 0.35
Identities = 17/48 (35%), Positives = 24/48 (49%)
Frame = -2
Query: 5 LLLLCFFIFVTFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNES 52
LLL+ F F TS PS+S+S+PT I L KP++ +
Sbjct: 252 LLLIVIFFFSPVLLSLAVTSTMPSTSTSNPTLICGTPLGDGVKPESSN 109
>TC216873 similar to UP|Q9SCQ0 (Q9SCQ0) RNA helicase-like protein
(At3g53110), partial (67%)
Length = 1840
Score = 29.6 bits (65), Expect = 1.0
Identities = 13/42 (30%), Positives = 23/42 (53%)
Frame = +1
Query: 115 PEDGFKLGENIYWGSGSDWTPTDAVKAWADEEKYYTYVTNSC 156
P G + + Y+G+G DW P D+ + + +E + V +SC
Sbjct: 658 PGCGSCIEKGTYYGTGGDWDPRDSQEVY*LQETWDHEVEDSC 783
>TC206927 homologue to UP|Q9ZPK4 (Q9ZPK4) Glutamyl-tRNA reductase, partial
(69%)
Length = 1499
Score = 29.3 bits (64), Expect = 1.3
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = -1
Query: 9 CFFIFV-TFTTKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQ-LCWNCMQ 66
CF ++ T T L +P S S SP + ++ Q+ P E + S LC++C
Sbjct: 953 CFLWYIITHLT*ALLKLSKPDSFSISPQFLNSRNSFQRVPPSFELLRFFSNDCLCFHCFS 774
Query: 67 ESLEFLFRHNLVR 79
++ F+ HNL +
Sbjct: 773 *AILFIGSHNLFK 735
>TC205186 similar to UP|Q8H9B8 (Q8H9B8) Low temperature and salt responsive
protein, partial (96%)
Length = 534
Score = 28.9 bits (63), Expect = 1.7
Identities = 13/34 (38%), Positives = 22/34 (64%)
Frame = +1
Query: 2 KSHLLLLCFFIFVTFTTKTLSTSPQPSSSSSSPT 35
+ +L +CF +F TTK +T+ P+S+SS P+
Sbjct: 46 REKILRICFLLFSF*TTKWPTTAQLPASTSSLPS 147
>TC214878 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit binding-protein
beta subunit, chloroplast precursor (60 kDa chaperonin
beta subunit) (CPN-60 beta), partial (26%)
Length = 687
Score = 28.5 bits (62), Expect = 2.2
Identities = 13/39 (33%), Positives = 19/39 (48%)
Frame = -3
Query: 48 PDNESIYKVSKQLCWNCMQESLEFLFRHNLVRASKWELP 86
PDN +I S ++C S+ FL L+ + W LP
Sbjct: 412 PDNLAILSFSFSFSYSCSAASIRFLICATLLFTASWVLP 296
>TC214210
Length = 959
Score = 28.5 bits (62), Expect = 2.2
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 10/65 (15%)
Frame = +3
Query: 15 TFTTKTLSTSPQPSSSSSSPTQIYN----------QYLSQQKKPDNESIYKVSKQLCWNC 64
+ T + + + S SS++PT+ N Q KKP+N++ +L WN
Sbjct: 282 SLTNQAVVSHAGSSKSSTNPTEFVNHGVILWNQTRQRWVGNKKPENQTQQLQEPKLSWNA 461
Query: 65 MQESL 69
ESL
Sbjct: 462 TYESL 476
>TC214876 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit binding-protein
beta subunit, chloroplast precursor (60 kDa chaperonin
beta subunit) (CPN-60 beta), partial (85%)
Length = 1797
Score = 28.5 bits (62), Expect = 2.2
Identities = 13/39 (33%), Positives = 19/39 (48%)
Frame = -1
Query: 48 PDNESIYKVSKQLCWNCMQESLEFLFRHNLVRASKWELP 86
PDN +I S ++C S+ FL L+ + W LP
Sbjct: 1005 PDNLAILSFSFSFSYSCSAASIRFLICATLLFTASWVLP 889
>TC228256 weakly similar to UP|Q9ATY4 (Q9ATY4) MAP kinase phosphatase
(Fragment), partial (10%)
Length = 1848
Score = 28.5 bits (62), Expect = 2.2
Identities = 14/40 (35%), Positives = 24/40 (60%)
Frame = +3
Query: 18 TKTLSTSPQPSSSSSSPTQIYNQYLSQQKKPDNESIYKVS 57
+ +LS+S SSSSSP+ + +S P ++S+ +VS
Sbjct: 390 SSSLSSSSSSVSSSSSPSYVSPDSISSDSSPHSKSLSEVS 509
>TC229330 similar to GB|AAQ56838.1|34098911|BT010395 At1g16900 {Arabidopsis
thaliana;} , partial (73%)
Length = 1436
Score = 28.5 bits (62), Expect = 2.2
Identities = 16/55 (29%), Positives = 28/55 (50%)
Frame = -1
Query: 26 QPSSSSSSPTQIYNQYLSQQKKPDNESIYKVSKQLCWNCMQESLEFLFRHNLVRA 80
QPS + + T+I LS++ KPD I + +L + + L +H++ RA
Sbjct: 212 QPSITPITATEIAAAGLSRRNKPDARDIAYIENELGKKLVLAKKQPLVKHSIARA 48
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.133 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,473,946
Number of Sequences: 63676
Number of extensions: 243643
Number of successful extensions: 1926
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 1836
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1890
length of query: 206
length of database: 12,639,632
effective HSP length: 93
effective length of query: 113
effective length of database: 6,717,764
effective search space: 759107332
effective search space used: 759107332
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC127169.12


