

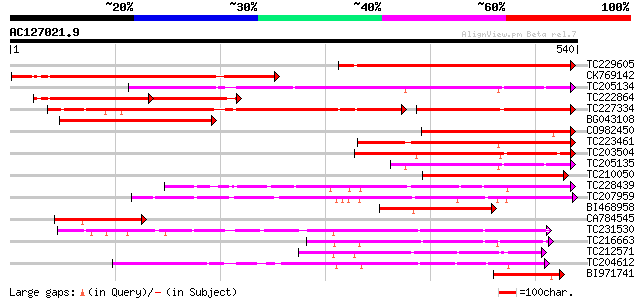
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127021.9 + phase: 0
(540 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229605 weakly similar to UP|Q8W2N1 (Q8W2N1) Cytochrome P450-de... 369 e-102
CK769142 335 3e-92
TC205134 similar to UP|C862_ARATH (O23066) Cytochrome P450 86A2 ... 318 5e-87
TC222864 similar to GB|AAO64841.1|29028924|BT005906 At5g63450 {A... 177 6e-70
TC227334 weakly similar to UP|Q9LXX7 (Q9LXX7) Cytochrome P450-li... 244 7e-65
BG043108 222 3e-58
CO982450 221 8e-58
TC223461 similar to UP|C941_VICSA (O81117) Cytochrome P450 94A1 ... 213 2e-55
TC203504 weakly similar to PIR|T00514|T00514 cytochrome P450 hom... 168 6e-42
TC205135 similar to UP|C862_ARATH (O23066) Cytochrome P450 86A2 ... 142 3e-34
TC210050 similar to PIR|T00864|T00864 cytochrome P450 homolog F1... 128 6e-30
TC228439 similar to UP|Q93VK5 (Q93VK5) At1g31800/68069_m00159, p... 118 6e-27
TC207959 UP|C972_SOYBN (O48921) Cytochrome P450 97B2 , complete 114 8e-26
BI468958 112 5e-25
CA784545 103 2e-22
TC231530 homologue to UP|C7DA_SOYBN (O48923) Cytochrome P450 71D... 100 2e-21
TC216663 UP|C823_SOYBN (O49858) Cytochrome P450 82A3 (P450 CP6)... 100 3e-21
TC212571 similar to UP|Q9FF18 (Q9FF18) Cytochrome P450-like prot... 99 4e-21
TC204612 UP|Q9SWR5 (Q9SWR5) Cytochrome P450 monooxygenase CYP93C... 93 3e-19
BI971741 92 4e-19
>TC229605 weakly similar to UP|Q8W2N1 (Q8W2N1) Cytochrome P450-dependent
fatty acid hydroxylase, partial (20%)
Length = 741
Score = 369 bits (947), Expect = e-102
Identities = 173/226 (76%), Positives = 203/226 (89%)
Frame = +1
Query: 314 ESRKDLLSRFMGALNSHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRV 373
E+R DLLSRFMG+++ DD YLRDIVVSFLLAGRDT+A+ LTGFF+LLSK+P+VEE I+
Sbjct: 13 EARNDLLSRFMGSID--DDVYLRDIVVSFLLAGRDTIAAGLTGFFMLLSKSPEVEELIKE 186
Query: 374 ELDRVMNPNQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKG 433
E RVM P QE +FEQ REMHYLN AIH+SMRLFPP+QFDSKFA EDDVLPDGTF++KG
Sbjct: 187 EAGRVMGPGQEFPSFEQIREMHYLNAAIHDSMRLFPPIQFDSKFATEDDVLPDGTFVRKG 366
Query: 434 SRVTYHPYAMGRMENIWGPDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAI 493
SRVTYHPYAMGRMENIWGPDCL+F+PERWL+DGVFVP+CPFKYPVFQAG RVCLGK+LA+
Sbjct: 367 SRVTYHPYAMGRMENIWGPDCLDFRPERWLRDGVFVPECPFKYPVFQAGVRVCLGKDLAL 546
Query: 494 VEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPVKIYER 539
+EMKSVV +LV+RFD+RV P+QEP+FAPGLTA+ RGG PV++ ER
Sbjct: 547 MEMKSVVVALVRRFDIRVAQPDQEPRFAPGLTATLRGGFPVRVCER 684
>CK769142
Length = 753
Score = 335 bits (859), Expect = 3e-92
Identities = 178/257 (69%), Positives = 202/257 (78%), Gaps = 1/257 (0%)
Frame = +2
Query: 2 PFPSFYISSYSQTAKRNIKQIEMSFYFHQDSLSLQSTMSTTFTFLFFSFTLLFSFFSFLL 61
P P F + T + I + EMSF Q LSLQS MS T FLFF+FTLLFSFFSFL+
Sbjct: 14 PSPHFLSFTPPPTPHKKI*R-EMSF---QGFLSLQS-MSATCGFLFFAFTLLFSFFSFLV 178
Query: 62 FISRIKPWCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQESPTGTIHVHVLGNTITSNPEN 121
F+SR+KPWCNC+ CK++LT+SW NKF NL D+YTHLL++SPTGTIHVHVL NTITSNP N
Sbjct: 179 FVSRMKPWCNCDVCKSYLTVSWLNKFPNLCDWYTHLLRKSPTGTIHVHVLENTITSNPVN 358
Query: 122 VEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVVIRSYAME 181
VEYILKTNF NYPKGK FS ILGDLLGRGIFNVDG SWK QRKMASLELGSV IR+ AME
Sbjct: 359 VEYILKTNFQNYPKGKPFSVILGDLLGRGIFNVDGESWKLQRKMASLELGSVAIRTNAME 538
Query: 182 LVIEEIKTRLLPLIASVAEKKTASKADNTSEDV-LLDMQDILRRFSFDNICKFSFGLDPC 240
LV EEI RL+P I + D ++ V +LD+QDILRRFSFDNICKFSFGLDP
Sbjct: 539 LVNEEIHARLIPFIMG------SVTHDELNDSVCVLDLQDILRRFSFDNICKFSFGLDPG 700
Query: 241 CLVPSLPVSNLANAFDL 257
CL+P+LPVSNLA+AFDL
Sbjct: 701 CLLPNLPVSNLADAFDL 751
>TC205134 similar to UP|C862_ARATH (O23066) Cytochrome P450 86A2 , partial
(87%)
Length = 2056
Score = 318 bits (814), Expect = 5e-87
Identities = 184/440 (41%), Positives = 267/440 (59%), Gaps = 14/440 (3%)
Frame = +2
Query: 114 TITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSV 173
T+T +P+N+E+ILK F+NYPKG + + DLLG GIFN DG +W FQRK A+LE +
Sbjct: 452 TVTCDPKNLEHILKLRFDNYPKGPTWQSAFHDLLGEGIFNSDGDTWLFQRKTAALEFTTR 631
Query: 174 VIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKF 233
+R V IK R P++A+ A+K+ S +D+QD+L R +FDNIC
Sbjct: 632 TLRQAMARWVNRAIKHRFCPILAT-AQKENKS----------VDLQDLLLRLTFDNICGL 778
Query: 234 SFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRFFNIGSEKKLKEAIK 293
+FG DP L LP + A +FD ++ + QR + ++WK+KR+ +G E L ++K
Sbjct: 779 AFGQDPQTLAVGLPDNAFALSFDRATEATLQRFILPE-ILWKLKRWLRLGMEVSLSRSLK 955
Query: 294 IVNDLANEMIKQRR-EIENGVESRKD-LLSRFMGALNSHDDEYLRDIVVSFLLAGRDTVA 351
+++ + +IK R+ E+ NG S D LLSRFM S+ +E+L+ + ++F+LAGRDT +
Sbjct: 956 HIDNYLSHIIKNRKLELLNGTGSHHDDLLSRFMRKKESYSEEFLQHVALNFILAGRDTSS 1135
Query: 352 SALTGFFILLSKNPKVEEKIRVEL---------DRVMNPNQECATFEQTREMHYLNGAIH 402
AL+ FF L KNP VEEKI EL D + +E FE+ + YL A+
Sbjct: 1136VALSWFFWLCIKNPHVEEKILHELCSVLKFTRGDDISTWLEEPLVFEEVDRLVYLKAALS 1315
Query: 403 ESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERW 462
E++RL+P V DSK ++DDVLP+GTF+ GS VTY Y++GRM+ IWG DCLEFKPERW
Sbjct: 1316ETLRLYPSVPEDSKHVVKDDVLPNGTFVPAGSAVTYSIYSVGRMKFIWGEDCLEFKPERW 1495
Query: 463 LK---DGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQ 519
L D + V + +K+ F AG R+CLGK+LA ++MKS+ A+++ R + V P +
Sbjct: 1496LSPEGDKIQV-QDSYKFVSFNAGPRLCLGKDLAYLQMKSIAAAVLLRHRL-AVAPGHRVE 1669
Query: 520 FAPGLTASFRGGLPVKIYER 539
LT + GL V +Y R
Sbjct: 1670QKMSLTLFMKYGLRVNVYPR 1729
>TC222864 similar to GB|AAO64841.1|29028924|BT005906 At5g63450 {Arabidopsis
thaliana;} , partial (11%)
Length = 570
Score = 177 bits (448), Expect(2) = 6e-70
Identities = 87/115 (75%), Positives = 97/115 (83%)
Frame = +3
Query: 23 EMSFYFHQDSLSLQSTMSTTFTFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMS 82
EMSF LSLQS MS T F FFSFTLLFSFFSFL+FISR+KPWCNC+ CK++LT+S
Sbjct: 9 EMSF---AGFLSLQS-MSATCGFFFFSFTLLFSFFSFLVFISRMKPWCNCDLCKSYLTVS 176
Query: 83 WSNKFVNLVDYYTHLLQESPTGTIHVHVLGNTITSNPENVEYILKTNFNNYPKGK 137
W KF NL D+YTHLL++SPTGTIHVHVLGNTITSNP NVE+ILKTNF NYPKGK
Sbjct: 177 WLKKFPNLCDWYTHLLRKSPTGTIHVHVLGNTITSNPHNVEHILKTNFQNYPKGK 341
Score = 106 bits (264), Expect(2) = 6e-70
Identities = 57/87 (65%), Positives = 65/87 (74%)
Frame = +1
Query: 134 PKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLP 193
P+ K FSTILGDLLGRGIFNVDG SWKFQRKMASLELGSV IR+YAMELV EEI RL+P
Sbjct: 331 PRVKPFSTILGDLLGRGIFNVDGESWKFQRKMASLELGSVAIRTYAMELVNEEIHARLIP 510
Query: 194 LIASVAEKKTASKADNTSEDVLLDMQD 220
++ S A + S +LD+QD
Sbjct: 511 IMESTARGELNSVC-------VLDLQD 570
>TC227334 weakly similar to UP|Q9LXX7 (Q9LXX7) Cytochrome P450-like protein,
partial (77%)
Length = 1754
Score = 244 bits (623), Expect = 7e-65
Identities = 141/350 (40%), Positives = 219/350 (62%), Gaps = 8/350 (2%)
Frame = +1
Query: 37 STMSTTFTFLFFSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVN----LVD 92
S+ +FLF S LLF + ++ F++R KP N K + + +F+ ++
Sbjct: 19 SSQMELLSFLFLS--LLFFLYLYIQFLARKKPRNN-KAFKDYPLIGTLPEFLKNRHRFLE 189
Query: 93 YYTHLLQESPTGT---IHVHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGR 149
+ T +L++SPT T + L +T+NP+NVE++LKT F+NYPKG++F +L D LG
Sbjct: 190 WTTQVLRDSPTNTGVFSRPYKLHGILTANPDNVEHVLKTKFDNYPKGERFIHLLQDFLGN 369
Query: 150 GIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADN 209
GIFN DG WK QRK AS E + +R++ ++ V E++TRLLP++ SKA
Sbjct: 370 GIFNSDGDLWKVQRKTASYEFSTKSLRNFVVDAVTFELQTRLLPIL---------SKASE 522
Query: 210 TSEDVLLDMQDILRRFSFDNICKFSFGLDPCCL-VPSLPVSNLANAFDLSSTLSAQRALT 268
T++ LD+QD+L RF+FDN+CK +F +DP CL AF+ ++ LS+ R ++
Sbjct: 523 TNKG--LDLQDLLERFAFDNVCKLAFNVDPACLGGDGTAGGEFMRAFEDAADLSSGRFMS 696
Query: 269 ASPLIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRREIENGVESRKDLLSRFMGALN 328
P +WK+K+ FN GSE++L+E+I V+ A+ +I+ R E ++ + +DLLSRF+ N
Sbjct: 697 ILPDVWKIKKLFNFGSERRLRESITTVHQFADSIIRSRLESKDKI-GDEDLLSRFIRTEN 873
Query: 329 SHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRV 378
+ E+LRD+V+SF+LAGRDT +SAL+ FF +LS P V+ KIR E++ V
Sbjct: 874 T-SPEFLRDVVISFILAGRDTTSSALSWFFWILSSRPDVQRKIRDEIETV 1020
Score = 174 bits (441), Expect = 9e-44
Identities = 81/152 (53%), Positives = 108/152 (70%)
Frame = +3
Query: 388 FEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRME 447
+E+ +EM YL AI E+MRL+PPV D+ L DDVLPDGT + KG VTYH YAMGRME
Sbjct: 1188 YEEVKEMRYLQAAISETMRLYPPVPVDTMECLNDDVLPDGTRVGKGWFVTYHTYAMGRME 1367
Query: 448 NIWGPDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRF 507
++WG DC EFKPERWL++ + PF+YPVF AG R+CLGKE+A ++MKS+ ASL++RF
Sbjct: 1368 SVWGKDCTEFKPERWLENRA---ESPFRYPVFHAGPRMCLGKEMAYIQMKSIAASLLERF 1538
Query: 508 DVRVVGPNQEPQFAPGLTASFRGGLPVKIYER 539
++ + + P+ LT +GGLPV + R
Sbjct: 1539 EIEALDKDTCPEHVLSLTMRIKGGLPVSVRVR 1634
>BG043108
Length = 451
Score = 222 bits (566), Expect = 3e-58
Identities = 107/150 (71%), Positives = 124/150 (82%)
Frame = +2
Query: 48 FSFTLLFSFFSFLLFISRIKPWCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQESPTGTIH 107
FSF LLFS FSFL+FISRIKPWCNC+ C ++LT++W NK NL +YT+LL SPTGTIH
Sbjct: 2 FSFPLLFSLFSFLIFISRIKPWCNCDLCTSYLTVTWLNKSPNLCHWYTNLLH*SPTGTIH 181
Query: 108 VHVLGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMAS 167
+HVLGNTITSN NVE+ILKTNF +YPKGK FSTILGDLLGRGI NV G SWK RKMAS
Sbjct: 182 LHVLGNTITSNSHNVEHILKTNFQDYPKGKPFSTILGDLLGRGIINVHGESWKLHRKMAS 361
Query: 168 LELGSVVIRSYAMELVIEEIKTRLLPLIAS 197
LEL V +R+YAMELV +EI TRL+P++ S
Sbjct: 362 LELCGVAMRTYAMELVNKEIHTRLIPIMES 451
>CO982450
Length = 754
Score = 221 bits (562), Expect = 8e-58
Identities = 100/149 (67%), Positives = 124/149 (83%), Gaps = 2/149 (1%)
Frame = -2
Query: 393 EMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGP 452
++HYL A HESMRLFPP+QFDSKF LEDDVLPDGT ++ G+RVTYHPYAMGR+E IWG
Sbjct: 753 QLHYLQAATHESMRLFPPIQFDSKFCLEDDVLPDGTKVESGTRVTYHPYAMGRLEEIWGC 574
Query: 453 DCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVV 512
DCLEF+P+RWLKDGVF P PF+YPVFQAG RVC+GKE+A++EMKSV SL+++F + ++
Sbjct: 573 DCLEFRPQRWLKDGVFQPMNPFEYPVFQAGLRVCVGKEVALMEMKSVAVSLLRKFHIELL 394
Query: 513 GPNQ--EPQFAPGLTASFRGGLPVKIYER 539
P P+F+PGLTA+FR GLPV + ER
Sbjct: 393 APLSFGNPRFSPGLTATFRFGLPVMVRER 307
>TC223461 similar to UP|C941_VICSA (O81117) Cytochrome P450 94A1
(P450-dependent fatty acid omega-hydroxylase) , partial
(35%)
Length = 720
Score = 213 bits (542), Expect = 2e-55
Identities = 108/214 (50%), Positives = 149/214 (69%), Gaps = 6/214 (2%)
Frame = +1
Query: 332 DEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQT 391
++++ DIV+SF+LAG+DT ++AL FF LLSKNP VE+++ E+ E +++
Sbjct: 10 EDFVTDIVISFILAGKDTTSAALMWFFWLLSKNPGVEKEVLKEIME----KPETPAYDEV 177
Query: 392 REMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWG 451
++M Y++ A+ ESMRL+PPV DSK A++DDVLPDGT +KKG+ VTYH YAMGRME+IWG
Sbjct: 178 KDMVYIHAALCESMRLYPPVSMDSKEAVDDDVLPDGTVVKKGTLVTYHVYAMGRMESIWG 357
Query: 452 PDCLEFKPERWLK-----DGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKR 506
D EFKPERWL+ FV + F YPVFQAG R+CLGKE+A ++MK +VA +++R
Sbjct: 358 EDWAEFKPERWLEKVETGKWKFVGRDSFTYPVFQAGPRICLGKEMAFMQMKRLVAGILRR 537
Query: 507 FD-VRVVGPNQEPQFAPGLTASFRGGLPVKIYER 539
F V + EP + LT+ GG PVKI +R
Sbjct: 538 FTVVPAMAKGVEPHYFAFLTSQMEGGFPVKIIDR 639
>TC203504 weakly similar to PIR|T00514|T00514 cytochrome P450 homolog
T20D16.19 - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (28%)
Length = 914
Score = 168 bits (425), Expect = 6e-42
Identities = 88/218 (40%), Positives = 135/218 (61%), Gaps = 7/218 (3%)
Frame = +3
Query: 329 SHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECA-- 386
+HDD++LRD V + +AGRDT+ SALT FF L++ NP VE KI E+ + ++
Sbjct: 27 AHDDKFLRDAVFNLFVAGRDTITSALTWFFWLVATNPSVEAKILEEMKEKLGTKEKSLGV 206
Query: 387 -TFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGR 445
+ E+ + + YL+GAI E++RLFPP+ F+ K A+ D+LP G + G+ + + YAMGR
Sbjct: 207 LSVEEVKRLVYLHGAICEALRLFPPIPFERKQAISSDMLPSGHRVNSGTMILFSLYAMGR 386
Query: 446 MENIWGPDCLEFKPERWLKDG---VFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVAS 502
E WG DC EFKPERW+ + V+VP +K+ F AG R CLGK+ + ++MK V +
Sbjct: 387 FEETWGKDCFEFKPERWISEKGGIVYVPS--YKFIAFNAGPRTCLGKDSSFIQMKMVATA 560
Query: 503 LVKRFDVRVV-GPNQEPQFAPGLTASFRGGLPVKIYER 539
++ ++ V+VV G P + + + GL V+I +R
Sbjct: 561 ILHKYRVQVVEGFVATPSLS--IVLLMKDGLKVQITKR 668
>TC205135 similar to UP|C862_ARATH (O23066) Cytochrome P450 86A2 , partial
(32%)
Length = 1096
Score = 142 bits (359), Expect = 3e-34
Identities = 81/189 (42%), Positives = 113/189 (58%), Gaps = 12/189 (6%)
Frame = +3
Query: 363 KNPKVEEKIRVEL---------DRVMNPNQECATFEQTREMHYLNGAIHESMRLFPPVQF 413
KNP+VEE I EL D + E F++ + YL A+ E++RL+P V
Sbjct: 3 KNPRVEENILNELCTVLLSTRGDNISTWLNEPLVFDEVDRLVYLKAALSETLRLYPSVPE 182
Query: 414 DSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWLK---DGVFVP 470
DSK ++DDVLP+GTF+ GS VTY Y++GRM+ IWG DCLEFKPERWL D + V
Sbjct: 183 DSKHVVKDDVLPNGTFVPAGSAVTYSIYSVGRMKFIWGEDCLEFKPERWLSPEGDKIQV- 359
Query: 471 KCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRG 530
+ +K+ F AG R+CLGK+LA ++MKS+ A+++ R + V P + LT +
Sbjct: 360 QDSYKFVSFNAGPRLCLGKDLAYLQMKSIAAAVLLRHRL-AVAPGHRVEQKMSLTLFMKY 536
Query: 531 GLPVKIYER 539
GL V +Y R
Sbjct: 537 GLKVNVYPR 563
>TC210050 similar to PIR|T00864|T00864 cytochrome P450 homolog F17K2.4 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(23%)
Length = 783
Score = 128 bits (322), Expect = 6e-30
Identities = 63/139 (45%), Positives = 85/139 (60%)
Frame = +2
Query: 394 MHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPD 453
MHYL+ A+ E++RL+P V D + A D+LPDG +KKG V Y Y MGRM +IWG D
Sbjct: 68 MHYLHAALTETLRLYPAVPADGRSAEAHDILPDGHKLKKGDGVYYLAYGMGRMCSIWGED 247
Query: 454 CLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVG 513
EF+PERWL +G+F P+ PFK+ F AG R+CLGK+ A +MK V +LV+ F ++
Sbjct: 248 AEEFRPERWLNNGIFQPESPFKFVAFHAGPRICLGKDFAYRQMKIVAMALVRFFRFKLSN 427
Query: 514 PNQEPQFAPGLTASFRGGL 532
Q + T GL
Sbjct: 428 RTQNVTYKVMFTLHIDKGL 484
>TC228439 similar to UP|Q93VK5 (Q93VK5) At1g31800/68069_m00159, partial (66%)
Length = 1543
Score = 118 bits (296), Expect = 6e-27
Identities = 111/405 (27%), Positives = 189/405 (46%), Gaps = 13/405 (3%)
Frame = +3
Query: 148 GRGIFNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKA 207
G+G+ DG W+ +R+ L + AM + + RL +K A+ +
Sbjct: 3 GKGLIPADGEIWRVRRRAIVPALHQKYVA--AMIGLFGQAADRL-------CQKLDAAAS 155
Query: 208 DNTSEDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRAL 267
D EDV +M+ + R + D I K F D L + + A + R++
Sbjct: 156 DG--EDV--EMESLFSRLTLDIIGKAVFNYDFDSLSND---TGIVEAVYTVLREAEDRSV 314
Query: 268 TASPLIWKMKRFFNIGSE-KKLKEAIKIVNDLANEMIK--QRREIENGVESRKDLLSR-- 322
P +W++ + +I +K+ A+K +ND +++I +R E ++ ++ ++
Sbjct: 315 APIP-VWEIPIWKDISPRLRKVNAALKFINDTLDDLIAICKRMVDEEELQFHEEYMNEQD 491
Query: 323 ---FMGALNSHDD---EYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELD 376
L S DD + LRD +++ L+AG +T A+ LT F LLSK P+V K++ E+D
Sbjct: 492 PSILHFLLASGDDVSSKQLRDDLMTMLIAGHETSAAVLTWTFYLLSKEPRVMSKLQEEVD 671
Query: 377 RVMNPNQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRV 436
V+ + T E +++ Y I+ES+RL+P + +LEDDVL + IK+G +
Sbjct: 672 SVL--GDQYPTIEDMKKLKYTTRVINESLRLYPQPPVLIRRSLEDDVLGEYP-IKRGEDI 842
Query: 437 TYHPYAMGRMENIWGPDCLEFKPERWLKDGVFVPKC--PFKYPVFQAGSRVCLGKELAIV 494
+ + R +W D +FKPERW DG + FKY F G R C+G A
Sbjct: 843 FISVWNLHRSPKLW-DDADKFKPERWALDGPSPNETNQNFKYLPFGGGPRKCVGDLFASY 1019
Query: 495 EMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPVKIYER 539
E +A L++RF+ ++ + G T GL + + R
Sbjct: 1020ETVVALAMLMRRFNFQIAVGAPPVEMTTGATIHTTQGLKMTVTHR 1154
>TC207959 UP|C972_SOYBN (O48921) Cytochrome P450 97B2 , complete
Length = 1863
Score = 114 bits (286), Expect = 8e-26
Identities = 113/467 (24%), Positives = 205/467 (43%), Gaps = 43/467 (9%)
Frame = +1
Query: 117 SNPENVEYILKTNFNNYPKGKQFSTILGDLLGRGIFNVDGHSWKFQRKMASLELGSVVIR 176
S+P +IL+ N +Y KG + IL ++G+G+ D +WK +R++ + + +
Sbjct: 415 SDPIVARHILRENAFSYDKGV-LADILEPIMGKGLIPADLDTWKQRRRVIAPAFHNSYLE 591
Query: 177 SYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLLDMQDILRRFSFDNICKFSFG 236
AM + R + + E + D+ + LD++ + D I F
Sbjct: 592 --AMVKIFTTCSERTILKFNKLLEGEGYDGPDS----IELDLEAEFSSLALDIIGLGVFN 753
Query: 237 LDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKMKRF-FNIGSEKKLKEAIKIV 295
D + PV + A+ T WK+ + + ++K ++ +K++
Sbjct: 754 YDFGSVTKESPVIKAV----YGTLFEAEHRSTFYIPYWKIPLARWIVPRQRKFQDDLKVI 921
Query: 296 NDLANEMIKQRREI--ENGVES--RKDLLS-------RFMGALNSHD--DEYLRDIVVSF 342
N + +I+ +E E VE ++D L+ RF+ + D D LRD +++
Sbjct: 922 NTCLDGLIRNAKESRQETDVEKLQQRDYLNLKDASLLRFLVDMRGADVDDRQLRDDLMTM 1101
Query: 343 LLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQTREMHYLNGAIH 402
L+AG +T A+ LT LL++NP +K + E+D V+ + TFE +E+ Y+ +
Sbjct: 1102LIAGHETTAAVLTWAVFLLAQNPSKMKKAQAEVDLVLGTGR--PTFESLKELQYIRLIVV 1275
Query: 403 ESMRLFPPVQFDSKFALEDDVLP-------DGTFIKKGSRVTYHPYAMGRMENIWG-PDC 454
E++RL+P + +L+ DVLP DG I G+ V Y + R W PD
Sbjct: 1276EALRLYPQPPLLIRRSLKSDVLPGGHKGEKDGYAIPAGTDVFISVYNLHRSPYFWDRPD- 1452
Query: 455 LEFKPERWL------------------KDGVFVPK---CPFKYPVFQAGSRVCLGKELAI 493
+F+PER+L G P F + F G R C+G + A+
Sbjct: 1453-DFEPERFLVQNKNEEIEGWAGLDPSRSPGALYPNEVISDFAFLPFGGGPRKCVGDQFAL 1629
Query: 494 VEMKSVVASLVKRFDVRVVGPNQEPQFAPGLTASFRGGLPVKIYERT 540
+E + L++ FDV + G + + G T + G+ ++ +R+
Sbjct: 1630MESTVALTMLLQNFDVELKGTPESVELVTGATIHTKNGMWCRLKKRS 1770
>BI468958
Length = 383
Score = 112 bits (279), Expect = 5e-25
Identities = 56/120 (46%), Positives = 77/120 (63%), Gaps = 9/120 (7%)
Frame = +3
Query: 353 ALTGFFILLSKNPKVEEKIRVELDRVMNPNQ---------ECATFEQTREMHYLNGAIHE 403
AL+ FF L+ +NPKVEEKI E+ ++ + E FE+ + YL A+ E
Sbjct: 12 ALSWFFWLVIQNPKVEEKILREICTILMETRGDDMAKWLDEPLDFEEVDRLVYLKAALSE 191
Query: 404 SMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPDCLEFKPERWL 463
++RL+P V DSK + DDVLPDGTF+ GS VTY Y+ GR+++ WG DC+EF+PERWL
Sbjct: 192 TLRLYPSVPEDSKHVVADDVLPDGTFVPAGSSVTYSIYSAGRLKSTWGEDCMEFRPERWL 371
>CA784545
Length = 433
Score = 103 bits (257), Expect = 2e-22
Identities = 48/90 (53%), Positives = 63/90 (69%), Gaps = 2/90 (2%)
Frame = +2
Query: 43 FTFLFFSFTLLFSFFSFLLFISRIKP--WCNCNTCKTFLTMSWSNKFVNLVDYYTHLLQE 100
F++L FS + F ++ I+R+ +C+C TC+T+LT SWS F NL D+Y HLL+
Sbjct: 164 FSYLLFSMITIILFLIIIIAIARLLKSFFCSCETCQTYLTSSWSETFDNLCDWYAHLLRN 343
Query: 101 SPTGTIHVHVLGNTITSNPENVEYILKTNF 130
SP TIH+HVL NTIT+N ENVEYILKT F
Sbjct: 344 SPNKTIHIHVLRNTITANAENVEYILKTKF 433
>TC231530 homologue to UP|C7DA_SOYBN (O48923) Cytochrome P450 71D10 ,
complete
Length = 1661
Score = 100 bits (248), Expect = 2e-21
Identities = 115/492 (23%), Positives = 214/492 (43%), Gaps = 21/492 (4%)
Frame = +1
Query: 46 LFFSFTLLFSFFSFLLFISRIKPWCNCNTCK------TFLTMSWSNKFVNL--VDYYTHL 97
++F ++LF FF F + R + +TCK T + ++ V V YY
Sbjct: 22 IYFITSILFIFFVFFKLVQRSDSKTS-STCKLPPGPRTLPLIGNIHQIVGSLPVHYYLKN 198
Query: 98 LQESPTGTIHVHV--LGNTITSNPENVEYILKTNFNNYPKGKQFSTILGDLL---GRGI- 151
L + +H+ + + N I ++PE + I+KT+ N+ F +L ++ G GI
Sbjct: 199 LADKYGPLMHLKLGEVSNIIVTSPEMAQEIMKTHDLNFSDRPDF--VLSRIVSYNGSGIV 372
Query: 152 FNVDGHSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTS 211
F+ G W+ RK+ ++EL + R + + EE L+ IA+ A ++ S
Sbjct: 373 FSQHGDYWRQLRKICTVELLTAK-RVQSFRSIREEEVAELVKKIAATASEEGGS------ 531
Query: 212 EDVLLDMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASP 271
+ ++ + +F + +FG + +SN+ L S +S
Sbjct: 532 ---IFNLTQSIYSMTFGIAARAAFGKKS--RYQQVFISNMHKQLMLLGGFSVADLYPSSR 696
Query: 272 LIWKMKRFFNIGSEKKLKEAIKIVNDLANEMIKQRR------EIENGVESRKDLLSRFMG 325
+ M G+ KL++ ++ + + ++I + + E VE D+L +F
Sbjct: 697 VFQMM------GATGKLEKVHRVTDRVLQDIIDEHKNRNRSSEEREAVEDLVDVLLKFQK 858
Query: 326 ALNSH-DDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQE 384
D+ ++ ++ + G +T +S + L +NP+V E+ + E+ RV +
Sbjct: 859 ESEFRLTDDNIKAVIQDIFIGGGETSSSVVEWGMSELIRNPRVMEEAQAEVRRVYDSKGH 1038
Query: 385 CATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMG 444
E ++ YL I E++RL PPV + +G I +RV + +A+G
Sbjct: 1039VDETE-LHQLIYLKSIIKETLRLHPPVPLLVPRVSRERCQINGYEIPSKTRVIINAWAIG 1215
Query: 445 RMENIWGPDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLV 504
R W + FKPER+L + F++ F AG R+C G AI ++ +A L+
Sbjct: 1216RNPKYWA-EAESFKPERFLNSSIDFRGTDFEFIPFGAGRRICPGITFAIPNIELPLAQLL 1392
Query: 505 KRFDVRVVGPNQ 516
FD ++ PN+
Sbjct: 1393YHFDWKL--PNK 1422
>TC216663 UP|C823_SOYBN (O49858) Cytochrome P450 82A3 (P450 CP6) , complete
Length = 1814
Score = 99.8 bits (247), Expect = 3e-21
Identities = 73/248 (29%), Positives = 126/248 (50%), Gaps = 12/248 (4%)
Frame = +1
Query: 283 GSEKKLKEAIKIVNDLANEMIKQRRE---IENGVESRKDLLSRFMGALNSHD------DE 333
G EK +K K ++ L +E +++ R+ + VES +D + + ALN D
Sbjct: 796 GHEKAMKANAKEIDKLLSEWLEEHRQKKLLGENVESDRDFMDVMISALNGAQIGAFDADT 975
Query: 334 YLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPNQECATFEQTRE 393
+ + +L G D+ A LT LL +NP K + E+D + ++ + ++
Sbjct: 976 ICKATSLELILGGTDSTAVTLTWALSLLLRNPLALGKAKEEIDMQIGKDEYIRESDISK- 1152
Query: 394 MHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYAMGRMENIWGPD 453
+ YL + E++RL+PP F S ++ + G IKKG+R+ ++ + + R ++W D
Sbjct: 1153 LVYLQAIVKETLRLYPPAPFSSPREFTENCILGGYHIKKGTRLIHNLWKIHRDPSVWS-D 1329
Query: 454 CLEFKPERWL---KDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVR 510
LEFKPER+L KD V + F+ F +G RVC G L + + +A+L+ FD
Sbjct: 1330 PLEFKPERFLTTHKD-VDLRGHNFELLPFGSGRRVCAGMSLGLNMVHFTLANLLHSFD-- 1500
Query: 511 VVGPNQEP 518
++ P+ EP
Sbjct: 1501 ILNPSAEP 1524
>TC212571 similar to UP|Q9FF18 (Q9FF18) Cytochrome P450-like protein, partial
(48%)
Length = 883
Score = 99.4 bits (246), Expect = 4e-21
Identities = 74/250 (29%), Positives = 119/250 (47%), Gaps = 14/250 (5%)
Frame = +2
Query: 276 MKRFFNIGSEKKLKEAIKIVNDLANEMIKQRR---EIENGVESRKDLLSRFMGAL----- 327
+ RFF +++K V L E+I+ R+ EI DLL + +
Sbjct: 20 LHRFFPSKYNREIKSLKMEVETLLMEIIQSRKDCVEIGRSNSYGNDLLGMLLNEMQKKKK 199
Query: 328 ------NSHDDEYLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNP 381
+S + + + D +F AG +T A LT +LL+ N ++K+R E+ V N
Sbjct: 200 GNGNNNSSINLQLVMDQCKTFFFAGHETTALLLTWTVMLLASNTSWQDKVRAEVKSVCNG 379
Query: 382 NQECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPY 441
+ +Q ++ L+ I+ESMRL+PP + ED VL D +I KG +
Sbjct: 380 G--IPSLDQLSKLTLLHMVINESMRLYPPASVLPRMVFEDIVLGD-LYIPKGLSIWIPVL 550
Query: 442 AMGRMENIWGPDCLEFKPERWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVA 501
A+ E +WG D EF PER+ FVP ++ F +G R C+G+ A++E K ++A
Sbjct: 551 AIHHSEKLWGKDANEFNPERFTSKS-FVPG---RFLPFASGPRNCVGQAFALMEAKIILA 718
Query: 502 SLVKRFDVRV 511
L+ RF +
Sbjct: 719 MLISRFSFTI 748
>TC204612 UP|Q9SWR5 (Q9SWR5) Cytochrome P450 monooxygenase CYP93C1v2p,
complete
Length = 1912
Score = 93.2 bits (230), Expect = 3e-19
Identities = 101/439 (23%), Positives = 192/439 (43%), Gaps = 23/439 (5%)
Frame = +3
Query: 99 QESPTGTIHVHVLGNTITSNPENVEYILKTN-FNNYPKGKQFSTILGDLLGRGIFNVD-G 156
+ P +++ + + S PE + L+T+ ++ Q S I + V G
Sbjct: 270 KHGPLFSLYFGSMPTVVASTPELFKLFLQTHEATSFNTRFQTSAIRRLTYDSSVAMVPFG 449
Query: 157 HSWKFQRKMASLELGSVVIRSYAMELVIEEIKTRLLPLIASVAEKKTASKADNTSEDVLL 216
WKF RK+ +L + + L ++I+ + L ++A AE + L
Sbjct: 450 PYWKFVRKLIMNDLLNATTVNKLRPLRTQQIR-KFLRVMAQGAEAQKP-----------L 593
Query: 217 DMQDILRRFSFDNICKFSFGLDPCCLVPSLPVSNLANAFDLSSTLSAQRALTASPLIWKM 276
D+ + L +++ I G + + ++A L + + IW +
Sbjct: 594 DLTEELLKWTNSTISMMMLG-------EAEEIRDIAR-----EVLKIFGEYSLTDFIWPL 737
Query: 277 KRFFNIGSEKKLKEAIKIVNDLANEMIKQRREI----ENGVESRKDLLSRFMGALNSHDD 332
K EK++ + + + + +IK+RREI +NG ++ F+ L +
Sbjct: 738 KHLKVGKYEKRIDDILNKFDPVVERVIKKRREIVRRRKNGEVVEGEVSGVFLDTLLEFAE 917
Query: 333 E----------YLRDIVVSFLLAGRDTVASALTGFFILLSKNPKVEEKIRVELDRVMNPN 382
+ +++ +VV F AG D+ A A L NPKV EK R E+ V+ +
Sbjct: 918 DETMEIKITKDHIKGLVVDFFSAGTDSTAVATEWALAELINNPKVLEKAREEVYSVVGKD 1097
Query: 383 QECATFEQTREMHYLNGAIHESMRLFPPVQFDSKFALEDDVLPDGTFIKKGSRVTYHPYA 442
+ + T+ + Y+ + E+ R+ PP+ + E+ + +G I +G+ + ++ +
Sbjct: 1098RLVDEVD-TQNLPYIRAIVKETFRMHPPLPVVKRKCTEECEI-NGYVIPEGALILFNVWQ 1271
Query: 443 MGRMENIWGPDCLEFKPERWLKDGVFVPKCP-------FKYPVFQAGSRVCLGKELAIVE 495
+GR W EF+PER+L+ G P F+ F +G R+C G LA
Sbjct: 1272VGRDPKYWDRPS-EFRPERFLETGAEGEAGPLDLRGQHFQLLPFGSGRRMCPGVNLATSG 1448
Query: 496 MKSVVASLVKRFDVRVVGP 514
M +++ASL++ FD++V+GP
Sbjct: 1449MATLLASLIQCFDLQVLGP 1505
>BI971741
Length = 427
Score = 92.4 bits (228), Expect = 4e-19
Identities = 42/74 (56%), Positives = 57/74 (76%), Gaps = 6/74 (8%)
Frame = -2
Query: 461 RWLKDGVFVPKCPFKYPVFQAGSRVCLGKELAIVEMKSVVASLVKRFDVRVVGP------ 514
RWLK+G+F P+ PFKYPVFQAG RVCLGKE+A++E+KSV SL++RF + + P
Sbjct: 426 RWLKEGLFCPQNPFKYPVFQAGIRVCLGKEMALLELKSVALSLLRRFHIELAAPPPHHHH 247
Query: 515 NQEPQFAPGLTASF 528
+ P+F+PGLTA+F
Sbjct: 246 HSTPRFSPGLTATF 205
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.323 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,427,346
Number of Sequences: 63676
Number of extensions: 366241
Number of successful extensions: 3267
Number of sequences better than 10.0: 200
Number of HSP's better than 10.0 without gapping: 3067
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3145
length of query: 540
length of database: 12,639,632
effective HSP length: 102
effective length of query: 438
effective length of database: 6,144,680
effective search space: 2691369840
effective search space used: 2691369840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC127021.9


