

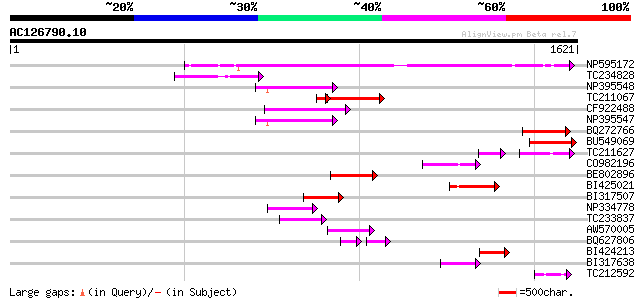
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126790.10 + phase: 0
(1621 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 706 0.0
TC234828 186 7e-47
NP395548 reverse transcriptase [Glycine max] 170 4e-42
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 142 5e-41
CF922488 160 3e-39
NP395547 reverse transcriptase [Glycine max] 155 1e-37
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 151 2e-36
BU549069 146 6e-35
TC211627 85 3e-30
CO982196 118 2e-26
BE802896 116 9e-26
BI425021 113 8e-25
BI317507 105 2e-22
NP334778 reverse transcriptase [Glycine max] 100 5e-21
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 93 1e-18
AW570005 89 2e-17
BQ627806 53 1e-15
BI424213 80 9e-15
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 76 1e-13
TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 75 2e-13
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 706 bits (1821), Expect = 0.0
Identities = 421/1142 (36%), Positives = 627/1142 (54%), Gaps = 28/1142 (2%)
Frame = +1
Query: 501 VLFDPGATYSFVSLYFAPRLGKSSSFLDE---TLVVTTPVGNNVLAKSVYYSCDVSIEGK 557
+L D G++ +F+ PR+ + E L V G + A+ + + I+G+
Sbjct: 1210 ILVDGGSSDNFIQ----PRVAQVLKLPVEPAPNLRVLVGNGQILSAEGIVQQLPLHIQGQ 1377
Query: 558 VLPADLVVLNMVDFDVILGMDWL-SLHHATVDCHNKVVKFKPPGEATFSFQGE-RSWVPN 615
+ + +L + DVILG WL +L D +KF + + QGE S
Sbjct: 1378 EVKVPVYLLQISGADVILGSTWLATLGPHVADYAALTLKFF-QNDKFITLQGEGNSEATQ 1554
Query: 616 NLISSLR--ANKLLSRGCQGYLALVRDVQAGEEKLENVP---------IVCEFSDVFPEE 664
+ R N C + ++V E+ L+++P ++ ++ VF
Sbjct: 1555 AQLHHFRRLQNTKSIEECFAIQLIQKEVP--EDTLKDLPTNIDPELAILLHTYAQVFAVP 1728
Query: 665 LPGIPPDREIEFSIDLIPGTQPISIPPYRMAPAELKELREQLQDLLDKGFIRASTSPWGA 724
+PP RE + +I L G+ P+ + PYR + ++ + +Q++L +G I+ S SP+
Sbjct: 1729 -ASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSL 1905
Query: 725 PVLFVKKKDGSMRLCVDYRQLNKVTIKNKYPLPRIDELFDQLQGAQCFSKIDLRSGYHQL 784
P+L VKKKDGS R C DYR LN +T+K+ +P+P +DEL D+L GAQ FSK+DLRSGYHQ+
Sbjct: 1906 PILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQI 2085
Query: 785 KIKSEDISKTAFRTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILI 844
++ ED KTAFRT +GHYE+LVM FGLTNAPA F LMN++F+ L +FV+VF DDILI
Sbjct: 2086 LVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILI 2265
Query: 845 YSKSREEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSKVEAV 904
YS S ++H +HL VLQTL++ QL+A+ SKC F V +LGH VS G+S++ +KV+AV
Sbjct: 2266 YSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAV 2445
Query: 905 QNWPRPTSVKEIRSFLGLAGYYRRFVKDFSKLAFPLTRLTQKKVEFKWTDACEESFQKLK 964
+WP P +VK++R FLGL GYYRRF+K ++ +A PLT L QK F W + E +F KLK
Sbjct: 2446 LDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAGPLTDLLQKD-SFLWNNEAEAAFVKLK 2622
Query: 965 ECLISAPILALPTSGGGYVVYCDASRVGLGCVLMQHGKVIAYASRQLKRHEQNYPTHDLE 1024
+ + AP+L+LP +++ DAS +G+G VL Q+G IAY S++L Q + E
Sbjct: 2623 KAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQNGHPIAYFSKKLAPRMQKQSAYTRE 2802
Query: 1025 MAAVIFALKIWRHYLYGETCEIYTDHKSLKYIFEQRDLNLRQRRWMELLKDYDCTILYHP 1084
+ A+ AL +RHYL G I TD +SLK + +Q Q+ W+ YD I Y P
Sbjct: 2803 LLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKP 2982
Query: 1085 GKANVVADALSRKSMGSLAHLTAIKRPIVKEFQEIVESGVQFEIDHSRTLLAHMKFRSTL 1144
GK N ADALSR M LA + S
Sbjct: 2983 GKDNQAADALSRMFM-----------------------------------LAWSEPHSIF 3057
Query: 1145 IDDIKQAQSQDSELMKMVDNVRNGKVSNFSVDSEGVLWLKSRICVPNVDDLRRKILEEAH 1204
+++++ D L ++++ + G ++ EG+L+ K R+ +P +++ KIL+E H
Sbjct: 3058 LEELRARLISDPHLKQLMETYKQGADASHYTVREGLLYWKDRVVIPAEEEIVNKILQEYH 3237
Query: 1205 HSSYTIHPGSNKMYQDLREFYWWEGMKRDVANFVSKCLVCQQVKAEHQKPAGLLQPIEIP 1264
S H G + L+ ++W M+ DV ++ KCL+CQQ K+ + PAGLLQP+ IP
Sbjct: 3238 SSPIGGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIP 3417
Query: 1265 KWKWEGIAMDFVTGLPRTQKGFDSVWVIIDRLTKSAHFLPVKTTYTASQYAKIYLEEIVS 1324
+ WE +AMDF+TGLP + G + V+IDRLTK AHF+P+K Y + A+ ++ IV
Sbjct: 3418 QQVWEDVAMDFITGLPNS-FGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVK 3594
Query: 1325 LHGVPISIISDRGAQFTAQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQILEDMLRAC 1384
LHG+P SI+SDR FT+ FW+ GT L +S+A+HPQ+DGQSE + LE LR
Sbjct: 3595 LHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCF 3774
Query: 1385 VLDLGGSWDRYLPMMEFAYNNSYQSSIQMAPFEALYGRR----CRSPIGWFEVGEAKLVG 1440
+ W + LP EF YN +Y S+ M PF ALYGR R + E +
Sbjct: 3775 TYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLTRQACSIDDPAEVR--- 3945
Query: 1441 PELIQDAIDKVKLIRDRLVTAQSRQKSYSDKRRRPLEFTVGEHVFLRVSPMK---GVLRF 1497
E + D + ++ L AQ K +DK+R + F +G+ V +++ P + VLR
Sbjct: 3946 -EQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLR- 4119
Query: 1498 GKKGKLTPRFIGPFEILERVGPVAYRLALPPDLSRVHPVFHISMLRKYIYDPSHVISHKD 1557
K KL+ R+ GPF++L ++G VAY+L L P +R+HPVFH+S L+ + + +D
Sbjct: 4120 -KNQKLSMRYFGPFKVLAKIGDVAYKLEL-PSAARIHPVFHVSQLKPFNG------TAQD 4275
Query: 1558 VQLDENLSYTE-----HPVALVDKGVRRLRSKDIVSVKVLWKGPSGEETTWEPEETIRGK 1612
L L+ TE PV ++ + I + V W+ +E TWE E I+
Sbjct: 4276 PYLPLPLTVTEMGPVMQPVKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKAS 4455
Query: 1613 YP 1614
YP
Sbjct: 4456 YP 4461
>TC234828
Length = 857
Score = 186 bits (472), Expect = 7e-47
Identities = 99/255 (38%), Positives = 151/255 (58%)
Frame = +3
Query: 471 ARVFALTRQDAQASNAVVTGTLSVCSQDAHVLFDPGATYSFVSLYFAPRLGKSSSFLDET 530
+RVFA++ +A AS+ ++ G + + VL+D GAT+SF+S RLG ++ L
Sbjct: 129 SRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYDSGATHSFISHACVERLGLCATELPYD 308
Query: 531 LVVTTPVGNNVLAKSVYYSCDVSIEGKVLPADLVVLNMVDFDVILGMDWLSLHHATVDCH 590
+VV+TP V V C + +EG+ ADL+ L + DVILGMDWLS +H +DC
Sbjct: 309 MVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLICLPLAHLDVILGMDWLSTNHIFLDCK 488
Query: 591 NKVVKFKPPGEATFSFQGERSWVPNNLISSLRANKLLSRGCQGYLALVRDVQAGEEKLEN 650
K++ F G+ VP+ + AN+ + + Y+ L + ++
Sbjct: 489 EKMLVFG--GDV----------VPSEPLKEDAANEE-TEDVRTYMVLFSMYVEEDAEVSC 629
Query: 651 VPIVCEFSDVFPEELPGIPPDREIEFSIDLIPGTQPISIPPYRMAPAELKELREQLQDLL 710
+P+V EF +VFP+++ +PP+RE+EF ID++PG P+SI PYRM+P EL E++ Q+QDLL
Sbjct: 630 IPVVSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPVELAEVKAQVQDLL 809
Query: 711 DKGFIRASTSPWGAP 725
K F+R S SPWGAP
Sbjct: 810 SKQFVRPSASPWGAP 854
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 170 bits (431), Expect = 4e-42
Identities = 92/254 (36%), Positives = 144/254 (56%), Gaps = 19/254 (7%)
Frame = +1
Query: 702 LREQLQDLLDKGFIRA-STSPWGAPVLFVKKKDG------------------SMRLCVDY 742
+R+++ LL+ G I S S W +PVL V KK+G S +LC+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 743 RQLNKVTIKNKYPLPRIDELFDQLQGAQCFSKIDLRSGYHQLKIKSEDISKTAFRTRYGH 802
R+LN+ T K+ +PLP +D++ ++L G + +D GY+Q+ + +D K AF +G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 803 YEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILIYSKSREEHEQHLRLVLQT 862
+ + + FGL NAP F M +F +++ + VF+DD ++ S E + L +VLQ
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 863 LREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSKVEAVQNWPRPTSVKEIRSFLGL 922
E L + KC F + LGH +S GI VD +K++ ++ P P++VK IRSFLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 923 AGYYRRFVKDFSKL 936
A +YRRF+KDF+K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 142 bits (359), Expect(2) = 5e-41
Identities = 76/164 (46%), Positives = 103/164 (62%)
Frame = +1
Query: 908 PRPTSVKEIRSFLGLAGYYRRFVKDFSKLAFPLTRLTQKKVEFKWTDACEESFQKLKECL 967
P SV +IRSF GLA +YRRFV +FS +A PL L +K + F W + E++F LKE L
Sbjct: 97 PTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKL 276
Query: 968 ISAPILALPTSGGGYVVYCDASRVGLGCVLMQHGKVIAYASRQLKRHEQNYPTHDLEMAA 1027
AP+LALP + + CDAS VG+ VL+Q G IAY S +L NYPT+D E+ A
Sbjct: 277 TKAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYA 456
Query: 1028 VIFALKIWRHYLYGETCEIYTDHKSLKYIFEQRDLNLRQRRWME 1071
+I A + W H+L + I++DH+SLKYI + LN R +W+E
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWVE 588
Score = 45.4 bits (106), Expect(2) = 5e-41
Identities = 17/40 (42%), Positives = 28/40 (69%)
Frame = +2
Query: 877 FWLESVAFLGHIVSKNGISVDPSKVEAVQNWPRPTSVKEI 916
F ++++ F G +V +NG+ +DP K++A+Q WP P V EI
Sbjct: 2 FCVDNIFFSGFVVGRNGVQMDPEKIKAIQEWPPP*KVWEI 121
>CF922488
Length = 741
Score = 160 bits (406), Expect = 3e-39
Identities = 90/245 (36%), Positives = 140/245 (56%)
Frame = +3
Query: 729 VKKKDGSMRLCVDYRQLNKVTIKNKYPLPRIDELFDQLQGAQCFSKIDLRSGYHQLKIKS 788
V K+DG + +CVDYR LN + K+K+PLP I+ L D FS +D SGY+Q+KI
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 789 EDISKTAFRTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILIYSKS 848
ED+ KT F T +G + + MSFGL N A + M +F + + + V++DD+++ S++
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 849 REEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSKVEAVQNWP 908
EEH +LR + + LR+ +L +KC F ++S L I S GI VD +KV+ +
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 909 RPTSVKEIRSFLGLAGYYRRFVKDFSKLAFPLTRLTQKKVEFKWTDACEESFQKLKECLI 968
+P + K+++ FLG Y RF+ PL L K KW C +F+++K+CLI
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 969 SAPIL 973
+ +L
Sbjct: 723 NPHVL 737
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 155 bits (393), Expect = 1e-37
Identities = 88/254 (34%), Positives = 133/254 (51%), Gaps = 19/254 (7%)
Frame = +1
Query: 702 LREQLQDLLDKGFIRA-STSPWGAPVLFVKKKDGSM------------------RLCVDY 742
+R+++ LL+ G I S S W +PV V KK G R+C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 743 RQLNKVTIKNKYPLPRIDELFDQLQGAQCFSKIDLRSGYHQLKIKSEDISKTAFRTRYGH 802
R+LN+ T K+ YPLP +D++ +L + +D SGY+Q+ + +D KTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 803 YEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILIYSKSREEHEQHLRLVLQT 862
+ + M FGL NA F M +F +++ + VF+DD + S +L VLQ
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 863 LREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSKVEAVQNWPRPTSVKEIRSFLGL 922
+ L + KC F ++ LGH +SK GI V K++ + P P +VK I SFLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 923 AGYYRRFVKDFSKL 936
G+YRRF+KDF+K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo}, partial
(9%)
Length = 410
Score = 151 bits (382), Expect = 2e-36
Identities = 75/136 (55%), Positives = 99/136 (72%)
Frame = -3
Query: 1467 SYSDKRRRPLEFTVGEHVFLRVSPMKGVLRFGKKGKLTPRFIGPFEILERVGPVAYRLAL 1526
SY DKRR+ LEF VG+HVFLRV+P GV R K KLTP FIGPF+IL++V VAY++AL
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 1527 PPDLSRVHPVFHISMLRKYIYDPSHVISHKDVQLDENLSYTEHPVALVDKGVRRLRSKDI 1586
PP L+ +H VFH+S + KYI DPSH+++ VQ+ ENL++ P+ + D + LR K+I
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 1587 VSVKVLWKGPSGEETT 1602
+ VKV+W G SGEE T
Sbjct: 48 LLVKVIWGGASGEEAT 1
>BU549069
Length = 615
Score = 146 bits (369), Expect = 6e-35
Identities = 71/135 (52%), Positives = 94/135 (69%)
Frame = -1
Query: 1486 LRVSPMKGVLRFGKKGKLTPRFIGPFEILERVGPVAYRLALPPDLSRVHPVFHISMLRKY 1545
L+V+P GV + K KLTP FIG F+IL+R PVAY++ALPP LS +H VFH+S LR Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 1546 IYDPSHVISHKDVQLDENLSYTEHPVALVDKGVRRLRSKDIVSVKVLWKGPSGEETTWEP 1605
I+DPSHV+ DVQ+ ENL+Y P+ + D+ + LR K+ VKV+W G SGE+ TWE
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 1606 EETIRGKYPHLFENQ 1620
E +R YP LFE++
Sbjct: 255 ESQMRVAYPSLFESE 211
>TC211627
Length = 1034
Score = 85.1 bits (209), Expect(2) = 3e-30
Identities = 54/161 (33%), Positives = 81/161 (49%), Gaps = 4/161 (2%)
Frame = +3
Query: 1457 RLVTAQSRQKSYSDKRRRPLEFTVGEHVFLRVSPMKGVLRFGKKGKLTPRFIGPFEILER 1516
RL Q K +D RR L F +G+ V++R+ P + KL+ RF GP++I R
Sbjct: 357 RLQKYQDSMKRIADSHRRDLTFNIGDWVYVRL*PYRQTSIQSTYTKLSKRFYGPYQIQAR 536
Query: 1517 VGPVAYRLALPPDLSRVHPVFHISMLRKYIYDPSHVISHKDVQLDENLSYTEHPVA---- 1572
VG VAYRL LPP S++HP+FH+S+L+ + H ++ S T HP+
Sbjct: 537 VGQVAYRLQLPP-TSKIHPIFHVSLLKVH-----HGPIPPELLALPPFSTTNHPLVQPLQ 698
Query: 1573 LVDKGVRRLRSKDIVSVKVLWKGPSGEETTWEPEETIRGKY 1613
+D + + I V V W + E+TTWE ++ Y
Sbjct: 699 FLDWKMDESTTPPIPQVLVQWTNLAPEDTTWESWTQLKDIY 821
Score = 67.0 bits (162), Expect(2) = 3e-30
Identities = 35/78 (44%), Positives = 44/78 (55%)
Frame = +2
Query: 1340 FTAQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQILEDMLRACVLDLGGSWDRYLPMM 1399
F + W GT+L STA+HPQTDGQ+E +ILE LRA V D W ++L +
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 1400 EFAYNNSYQSSIQMAPFE 1417
E YN S S I +PFE
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
>CO982196
Length = 812
Score = 118 bits (295), Expect = 2e-26
Identities = 60/166 (36%), Positives = 96/166 (57%)
Frame = +1
Query: 1179 GVLWLKSRICVPNVDDLRRKILEEAHHSSYTIHPGSNKMYQDLREFYWWEGMKRDVANFV 1238
G L+ K R+ + +L+E S H G + ++ + +W+GMK+ ++V
Sbjct: 316 GKLYFKDRLVLSKNSTKIPLLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYV 495
Query: 1239 SKCLVCQQVKAEHQKPAGLLQPIEIPKWKWEGIAMDFVTGLPRTQKGFDSVWVIIDRLTK 1298
+ C +C++ K PAGLL + IP W I+MDF+ GLP+ Q G D++ V++DRLTK
Sbjct: 496 AACEICRRNKTSTLSPAGLL*LLPIPTKVWTDISMDFIGGLPKAQ-GKDNILVVVDRLTK 672
Query: 1299 SAHFLPVKTTYTASQYAKIYLEEIVSLHGVPISIISDRGAQFTAQF 1344
AHF + YTA + A+++++E+V LHG P SI+SD F + F
Sbjct: 673 YAHFFALSHPYTAKEVAELFIKELVRLHGFPASIVSDXXRLFMSLF 810
>BE802896
Length = 416
Score = 116 bits (290), Expect = 9e-26
Identities = 61/137 (44%), Positives = 84/137 (60%), Gaps = 4/137 (2%)
Frame = -2
Query: 918 SFLGLAGYYRRFVKDFSKLAFPLTRLTQKKVEFKWTDACEESFQKLKECLISAPILALPT 977
SFLG AG+YRRF++DF K+A PL+ L QK+VEF + D C+ +F LK LI+ PI+ P
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 978 SGGGYVVYCDASRVGLGCVLMQH----GKVIAYASRQLKRHEQNYPTHDLEMAAVIFALK 1033
+ + CDAS LG VL Q +VI Y+SR L + NY T + E+ A++FAL+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 1034 IWRHYLYGETCEIYTDH 1050
+ YL G +Y DH
Sbjct: 55 KFHSYLLGTRIIVYIDH 5
>BI425021
Length = 426
Score = 113 bits (282), Expect = 8e-25
Identities = 54/143 (37%), Positives = 88/143 (60%)
Frame = -1
Query: 1258 LQPIEIPKWKWEGIAMDFVTGLPRTQKGFDSVWVIIDRLTKSAHFLPVKTTYTASQYAKI 1317
L P+ +P+ WE ++MDF+ GLP G +++V+++R +K H + T++TA A +
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLP-PYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASL 250
Query: 1318 YLEEIVSLHGVPISIISDRGAQFTAQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQIL 1377
+L ++ LHG P SI+SDR F + FW+ GT L +S+A+HPQTDGQ+E +++
Sbjct: 249 FLNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVI 70
Query: 1378 EDMLRACVLDLGGSWDRYLPMME 1400
E LRA V + R++P +E
Sbjct: 69 EQYLRAFVHGRPRNLGRFIPWVE 1
>BI317507
Length = 359
Score = 105 bits (261), Expect = 2e-22
Identities = 54/115 (46%), Positives = 77/115 (66%), Gaps = 1/115 (0%)
Frame = -1
Query: 841 DILIYSKSREEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGHIVSKNGISVDPSK 900
+ILIYS + H HL VL L++++L A KC F ++ +LGH++SK+ +++D +K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 901 VEAVQNWPRPTSVKEIRSFLGLAGYYRRFVKDFSKLA-FPLTRLTQKKVEFKWTD 954
V++V WP P +VK + SFL L GYYR+F+KD+ KLA PLT LT K FKW D
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLT-KNDGFKWGD 12
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 100 bits (249), Expect = 5e-21
Identities = 53/143 (37%), Positives = 84/143 (58%)
Frame = +3
Query: 737 RLCVDYRQLNKVTIKNKYPLPRIDELFDQLQGAQCFSKIDLRSGYHQLKIKSEDISKTAF 796
R+CVDYR LN+ + K+ +PLP ID L + FS +D SGY+Q+K+ ED+ KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 797 RTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFKPFLDRFVIVFIDDILIYSKSREEHEQHL 856
T +G + + VMSFGL N A + M +F+ + + + ++D+++ S+ EEH +L
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 857 RLVLQTLREKQLYAKFSKCEFWL 879
+ + LR+ +L KC F L
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 92.8 bits (229), Expect = 1e-18
Identities = 49/133 (36%), Positives = 80/133 (59%)
Frame = +2
Query: 772 FSKIDLRSGYHQLKIKSEDISKTAFRTRYGHYEFLVMSFGLTNAPAAFMDLMNRVFKPFL 831
FS +D SGY+Q+ + ED+ KT F T +G + + VM+FGL N A + M +F +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 832 DRFVIVFIDDILIYSKSREEHEQHLRLVLQTLREKQLYAKFSKCEFWLESVAFLGHIVSK 891
+ + V++DD++ S++ EH +L + L++ QL +KC F ++S LG IVS+
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 892 NGISVDPSKVEAV 904
GI +DP KV+A+
Sbjct: 362 KGIEIDPEKVKAL 400
>AW570005
Length = 413
Score = 88.6 bits (218), Expect = 2e-17
Identities = 49/134 (36%), Positives = 73/134 (53%)
Frame = -2
Query: 908 PRPTSVKEIRSFLGLAGYYRRFVKDFSKLAFPLTRLTQKKVEFKWTDACEESFQKLKECL 967
P P + + +R FL L G+YRRF+K ++ +A PL+ L K F W+ + +FQ LK +
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKD-SFVWSPEADVAFQALKNVV 236
Query: 968 ISAPILALPTSGGGYVVYCDASRVGLGCVLMQHGKVIAYASRQLKRHEQNYPTHDLEMAA 1027
+ +LALP + V DAS +G VL Q G IA+ S++ T+ E+AA
Sbjct: 235 TNTLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAA 56
Query: 1028 VIFALKIWRHYLYG 1041
+ +K WR YL G
Sbjct: 55 ITNVVKKWRQYLLG 14
>BQ627806
Length = 435
Score = 52.8 bits (125), Expect(2) = 1e-15
Identities = 28/68 (41%), Positives = 38/68 (55%)
Frame = +1
Query: 1020 THDLEMAAVIFALKIWRHYLYGETCEIYTDHKSLKYIFEQRDLNLRQRRWMELLKDYDCT 1079
T+ E+AA+ A+K WR YL G I TDH+SLK + Q Q+ ++ L +D T
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDYT 411
Query: 1080 ILYHPGKA 1087
I Y GKA
Sbjct: 412 IQYRAGKA 435
Score = 50.4 bits (119), Expect(2) = 1e-15
Identities = 22/60 (36%), Positives = 35/60 (57%)
Frame = +3
Query: 947 KVEFKWTDACEESFQKLKECLISAPILALPTSGGGYVVYCDASRVGLGCVLMQHGKVIAY 1006
K +F W + + +F +LK L AP+L LP +VV DAS +G+G +L Q+ +A+
Sbjct: 12 KDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLAF 191
>BI424213
Length = 426
Score = 79.7 bits (195), Expect = 9e-15
Identities = 38/87 (43%), Positives = 53/87 (60%)
Frame = +1
Query: 1342 AQFWKSFQAALGTRLNLSTAFHPQTDGQSERTIQILEDMLRACVLDLGGSWDRYLPMMEF 1401
+ FWK+ A LGT+L ST HPQTDGQ++ + L +LRA + SWD YLP +EF
Sbjct: 4 SHFWKTLWAKLGTKLLFSTTCHPQTDGQTKVVNRSLSTLLRALLKGNHKSWDEYLPHVEF 183
Query: 1402 AYNNSYQSSIQMAPFEALYGRRCRSPI 1428
AYN + + +PFE +YG +P+
Sbjct: 184 AYNRGVHRTTKQSPFEVVYGFNPLTPL 264
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 76.3 bits (186), Expect = 1e-13
Identities = 35/114 (30%), Positives = 64/114 (55%)
Frame = -2
Query: 1233 DVANFVSKCLVCQQVKAEHQKPAGLLQPIEIPKWKWEGIAMDFVTGLPRTQKGFDSVWVI 1292
+ A + CL CQ K E ++ LL P+ +P WE +++DF+TGL ++ V+
Sbjct: 350 ECAQMLPNCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGL-LPYHVHTAILVV 174
Query: 1293 IDRLTKSAHFLPVKTTYTASQYAKIYLEEIVSLHGVPISIISDRGAQFTAQFWK 1346
+D +K H + +++TA A ++++ + LHG+P S++SD F + FW+
Sbjct: 173 VDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQ 12
>TC212592 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 664
Score = 75.1 bits (183), Expect = 2e-13
Identities = 42/104 (40%), Positives = 62/104 (59%)
Frame = +2
Query: 1501 GKLTPRFIGPFEILERVGPVAYRLALPPDLSRVHPVFHISMLRKYIYDPSHVISHKDVQL 1560
GKL+ RF GPF+++ERVG VAYRL LP D +++HPVFH S+L+ + +P + L
Sbjct: 71 GKLSKRFFGPFQVVERVGKVAYRLQLPVD-AKLHPVFHCSLLKPFQGNPPDTAAPLPPTL 247
Query: 1561 DENLSYTEHPVALVDKGVRRLRSKDIVSVKVLWKGPSGEETTWE 1604
++ S V L + V + D + V V W+G S ++ TWE
Sbjct: 248 FDHQSVIAPLVILATRTV----NDDDIEVLVQWQGLSPDDATWE 367
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,237,836
Number of Sequences: 63676
Number of extensions: 990830
Number of successful extensions: 5954
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 5606
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5873
length of query: 1621
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1511
effective length of database: 5,635,272
effective search space: 8514895992
effective search space used: 8514895992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC126790.10


