

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126787.10 - phase: 0
(306 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
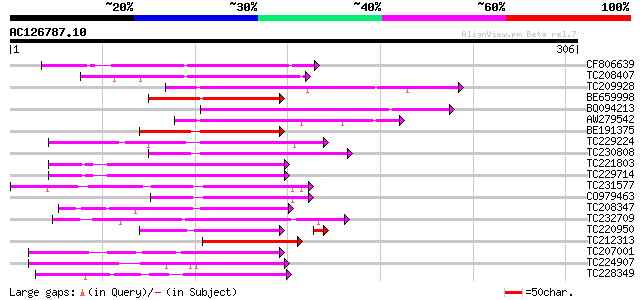
Score E
Sequences producing significant alignments: (bits) Value
CF806639 100 8e-22
TC208407 similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc finge... 97 7e-21
TC209928 weakly similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zin... 97 1e-20
BE659998 87 1e-17
BQ094213 similar to GP|28558782|gb| RING/C3HC4/PHD zinc finger-l... 84 1e-16
AW279542 similar to GP|28558782|gb| RING/C3HC4/PHD zinc finger-l... 83 1e-16
BE191375 83 1e-16
TC229224 weakly similar to UP|Q9LUR1 (Q9LUR1) RING zinc finger p... 81 5e-16
TC230808 similar to UP|Q9FFJ7 (Q9FFJ7) Similarity to C3HC4-type ... 78 4e-15
TC221803 weakly similar to UP|Q9SRM0 (Q9SRM0) F9F8.8 protein (RI... 77 7e-15
TC229714 weakly similar to UP|Q9SRM0 (Q9SRM0) F9F8.8 protein (RI... 75 5e-14
TC231577 weakly similar to UP|O23446 (O23446) Regulatory protein... 74 1e-13
CO979463 73 2e-13
TC208347 similar to UP|Q9XF63 (Q9XF63) RING-H2 zinc finger prote... 71 7e-13
TC232709 weakly similar to UP|Q9LZJ6 (Q9LZJ6) RING-H2 zinc finge... 70 9e-13
TC220950 similar to UP|Q9LS98 (Q9LS98) Similarity to RING zinc f... 68 1e-12
TC212313 68 6e-12
TC207001 similar to PIR|F86340|F86340 protein F2D10.34 [imported... 67 1e-11
TC224907 weakly similar to UP|Q6Z8A6 (Q6Z8A6) Zinc finger (C3HC4... 67 1e-11
TC228349 67 1e-11
>CF806639
Length = 708
Score = 100 bits (249), Expect = 8e-22
Identities = 59/150 (39%), Positives = 81/150 (53%)
Frame = +1
Query: 18 TPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRSPSRGL 77
+P + + V L +AF +YV ++C A +T+ S + + R +RGL
Sbjct: 121 SPSIAIIIVILVAALFLMAFFSIYV-RHC--------ADSPSTTVSPLTTARSRRAARGL 273
Query: 78 DNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLW 137
D +++ FP YS VK + K +LECA+CL EF+D LRL+ C HVFH ECID W
Sbjct: 274 DPAVIQTFPILEYSEVK-IHKIGKEALECAVCLCEFEDTETLRLIPKCDHVFHPECIDEW 450
Query: 138 LESHKTCPVCRTDLDSPPNQMSKHGEGNHN 167
L SH TCPVCR +L P + GN N
Sbjct: 451 LGSHTTCPVCRANL-VPTDSEDAIANGNAN 537
>TC208407 similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc finger-like
protein, partial (36%)
Length = 644
Score = 97.4 bits (241), Expect = 7e-21
Identities = 55/130 (42%), Positives = 74/130 (56%), Gaps = 6/130 (4%)
Frame = +2
Query: 39 VVYVCKYCFAGFFHTWA--LQRTTSGSLVRLSP----DRSPSRGLDNTLLEKFPTFLYSS 92
V+ + GFF + + S S+ L+ R +RGL+ +++ FPT YS+
Sbjct: 95 VILIAALFLMGFFSIYIRHCSDSPSASIRNLAAATGRSRRGTRGLEQAVIDTFPTLEYSA 274
Query: 93 VKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCRTDLD 152
VK + K +LECA+CL EF+D LRL+ C HVFH ECID WL SH TCPVCR +L
Sbjct: 275 VK-IHKLGKGTLECAVCLNEFEDTETLRLIPKCDHVFHPECIDEWLASHTTCPVCRANLV 451
Query: 153 SPPNQMSKHG 162
P + S HG
Sbjct: 452 PQPGE-SVHG 478
>TC209928 weakly similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc
finger-like protein, partial (18%)
Length = 1018
Score = 96.7 bits (239), Expect = 1e-20
Identities = 63/189 (33%), Positives = 85/189 (44%), Gaps = 28/189 (14%)
Frame = +1
Query: 85 FPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTC 144
FPT YS++KDL+K +LECA+CL +F D LRLL C HVFH CID WL H TC
Sbjct: 4 FPTLFYSNIKDLKKGNE-TLECAVCLTDFTDKDALRLLPKCNHVFHPHCIDSWLACHVTC 180
Query: 145 PVCRTDLDSPPNQMS------KHGEGNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEESDN 198
PVCR +L + +S EG+ N N E TS + + SD
Sbjct: 181 PVCRANLSQESSHVSITVPPHNEEEGSRNTTTNEATQIEQSTSNDVGQVCLGDPTPTSD- 357
Query: 199 VGEITRAQVHEGDQH----------------------DHHVGMSMQQQEDHRFSRSHSTG 236
+I + QH +++ G + + SRS+STG
Sbjct: 358 AAKIMYISEEQQQQHSSSEPTFEVELDPNTNSTTTTINNNGGDGVVVVSERNLSRSNSTG 537
Query: 237 HSIVTIRGE 245
H IV +G+
Sbjct: 538 HCIVEEQGK 564
>BE659998
Length = 868
Score = 86.7 bits (213), Expect = 1e-17
Identities = 37/73 (50%), Positives = 51/73 (69%)
Frame = -2
Query: 76 GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECID 135
GLD T+++ P LY++ D R ++S + +CA+CLLEF+DD +R L IC H FH +CID
Sbjct: 237 GLDETVIKTIPFSLYTAKYDARFDESRN-DCAVCLLEFEDDDYVRTLPICSHTFHVDCID 61
Query: 136 LWLESHKTCPVCR 148
WL SH CP+CR
Sbjct: 60 AWLRSHANCPLCR 22
>BQ094213 similar to GP|28558782|gb| RING/C3HC4/PHD zinc finger-like protein
{Cucumis melo}, partial (18%)
Length = 421
Score = 83.6 bits (205), Expect = 1e-16
Identities = 51/137 (37%), Positives = 68/137 (49%)
Frame = +1
Query: 104 LECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCRTDLDSPPNQMSKHGE 163
LECA+CL EF DD LRL+ CCHVFH +CID WL +H TCPVCR +L P E
Sbjct: 1 LECAVCLNEFRDDETLRLIPKCCHVFHSDCIDAWLANHSTCPVCRANLAPKPEDAPSSVE 180
Query: 164 GNHNNNNNNLNVQEGMTSLPCDDIHIDVRGEESDNVGEITRAQVHEGDQHDHHVGMSMQQ 223
++ + E + + GE++ V E R + + ++ S
Sbjct: 181 IQLSDPARPIGPNEPGHDPNYINPVEEREGEQNRIVTEPPRV-LDDPNRARPVRSKSTGF 357
Query: 224 QEDHRFSRSHSTGHSIV 240
F RSHSTGHS+V
Sbjct: 358 GIARLFPRSHSTGHSLV 408
>AW279542 similar to GP|28558782|gb| RING/C3HC4/PHD zinc finger-like protein
{Cucumis melo}, partial (12%)
Length = 434
Score = 83.2 bits (204), Expect = 1e-16
Identities = 50/132 (37%), Positives = 68/132 (50%), Gaps = 8/132 (6%)
Frame = +3
Query: 90 YSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCRT 149
YS++KDL K +LECA+CL +F LRLL C HVFH CID WL SH TCPVCR
Sbjct: 3 YSNIKDLNKANQ-TLECAVCLTDFTHKDSLRLLPKCNHVFHPHCIDSWLTSHVTCPVCRA 179
Query: 150 DLDSPPN-----QMSKHG-EGNHNNNNNNLNVQEG--MTSLPCDDIHIDVRGEESDNVGE 201
+L + + HG EG+ N E + P +DI+ D G+ + + +
Sbjct: 180 NLSQESSCHVSITVPPHGEEGSLGNMTTTTTTTEATQVEQRPSNDINQDCVGDPT-STSD 356
Query: 202 ITRAQVHEGDQH 213
T+ +QH
Sbjct: 357 TTKIIYISEEQH 392
>BE191375
Length = 495
Score = 83.2 bits (204), Expect = 1e-16
Identities = 37/78 (47%), Positives = 51/78 (64%)
Frame = -2
Query: 71 RSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFH 130
RS G+D ++E P F +SS+K ++ LECA+CL +F+D +LRLL C H FH
Sbjct: 413 RSRFSGIDKNVIESLPFFRFSSLKGSKE----GLECAVCLSKFEDVEILRLLPKCKHAFH 246
Query: 131 QECIDLWLESHKTCPVCR 148
+CID WLE H +CP+CR
Sbjct: 245 IDCIDHWLEKHSSCPICR 192
>TC229224 weakly similar to UP|Q9LUR1 (Q9LUR1) RING zinc finger protein-like,
partial (38%)
Length = 937
Score = 81.3 bits (199), Expect = 5e-16
Identities = 52/164 (31%), Positives = 78/164 (46%), Gaps = 13/164 (7%)
Frame = +1
Query: 22 LVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRSP---SRGLD 78
++ F + VL+LC ++ Y+ + L+RT LV + D +P SRGLD
Sbjct: 136 ILLFFVVVLMLCLHIYARWYLRRARRRQLRRQRELRRT---QLVFYNDDATPAAVSRGLD 306
Query: 79 NTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWL 138
+L P F + K ECA+CL EF+ R+L C H FH ECID+W
Sbjct: 307 AAILATLPVFTFDPEKT-------GPECAVCLSEFEPGETGRVLPKCNHSFHIECIDMWF 465
Query: 139 ESHKTCPVCRTDLD----------SPPNQMSKHGEGNHNNNNNN 172
SH TCP+CR ++ + P+ +S+ G G + N +
Sbjct: 466 HSHDTCPLCRAPVERAPEPEVVVITVPDPVSETGSGENENRTGS 597
>TC230808 similar to UP|Q9FFJ7 (Q9FFJ7) Similarity to C3HC4-type RING zinc
finger protein, partial (28%)
Length = 896
Score = 78.2 bits (191), Expect = 4e-15
Identities = 35/110 (31%), Positives = 61/110 (54%)
Frame = +1
Query: 76 GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECID 135
G+D +++E P F + +++ ++ L+CA+CL +F+ +LRLL C H FH EC+D
Sbjct: 10 GIDRSVVESLPVFRFGALRGQKE----GLDCAVCLNKFEAAEVLRLLPKCKHAFHVECVD 177
Query: 136 LWLESHKTCPVCRTDLDSPPNQMSKHGEGNHNNNNNNLNVQEGMTSLPCD 185
WL++H TCP+CR +D + + + ++ N +E L D
Sbjct: 178 TWLDAHSTCPLCRYRVDPEDILLVEDAKPFRQSHQQQRNKEEERVRLNLD 327
>TC221803 weakly similar to UP|Q9SRM0 (Q9SRM0) F9F8.8 protein (RING zinc
finger protein; 17145-17621), partial (34%)
Length = 772
Score = 77.4 bits (189), Expect = 7e-15
Identities = 40/130 (30%), Positives = 65/130 (49%)
Frame = +3
Query: 22 LVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRSPSRGLDNTL 81
+V F++ +L+ ++ +VC+Y L T + +P +P +GLD
Sbjct: 237 IVLFSIILLVTVLFIYTR-WVCRY-------QGRLPTTAFSAAAVHAPPLAPPQGLDPAS 392
Query: 82 LEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESH 141
++K P L+ + D + EC ICL EF D +++L C H FH +C+D WL H
Sbjct: 393 IKKLPIILHHAPADRDESAWDETECCICLGEFRDGEKVKVLPACDHYFHCDCVDKWLTHH 572
Query: 142 KTCPVCRTDL 151
+CP+CR L
Sbjct: 573 SSCPLCRASL 602
>TC229714 weakly similar to UP|Q9SRM0 (Q9SRM0) F9F8.8 protein (RING zinc
finger protein; 17145-17621), partial (34%)
Length = 915
Score = 74.7 bits (182), Expect = 5e-14
Identities = 38/130 (29%), Positives = 64/130 (49%)
Frame = +2
Query: 22 LVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRSPSRGLDNTL 81
+V F++ +L+ ++ +VC+Y L T + +P + +G+D
Sbjct: 227 IVLFSIILLVTVLFIYTR-WVCRY-------QGRLPTTAFTAAAAHAPPLAQPQGMDPAS 382
Query: 82 LEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESH 141
++K P L+ + D + EC ICL EF D +++L C H FH +C+D WL H
Sbjct: 383 IKKLPIILHHAPSDREESAWDETECCICLGEFRDGEKVKVLPACDHYFHCDCVDKWLTHH 562
Query: 142 KTCPVCRTDL 151
+CP+CR L
Sbjct: 563 SSCPLCRASL 592
>TC231577 weakly similar to UP|O23446 (O23446) Regulatory protein, partial
(15%)
Length = 779
Score = 73.6 bits (179), Expect = 1e-13
Identities = 52/175 (29%), Positives = 82/175 (46%), Gaps = 11/175 (6%)
Frame = +1
Query: 1 MSSSEPKYDTPPAPTYKTP--PALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQR 58
M++ EP ++ A K A++ F L ++I+ F F ++C ++
Sbjct: 106 MTTMEPDVESKYAHNGKVMVGTAILLFILIIIIIFFHTF-----VRFCRRRRRRLFSRSL 270
Query: 59 TTSGSLVRLSPDRSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSM 118
T+ S D LD ++++ PTF +S+ + +CA+CL EF D
Sbjct: 271 PTNSSAAASLDDPC----LDPSVIKSLPTFTFSAATHRSLQ-----DCAVCLSEFADGDE 423
Query: 119 LRLLTICCHVFHQECIDLWLESHKTCPVCRTDL-------DSPPN--QMSKHGEG 164
R+L C H FH CID W+ SH TCP+CRT + D+ P +S+ GEG
Sbjct: 424 GRVLPNCKHAFHAHCIDTWIGSHSTCPLCRTPVKPVTGSSDTEPGSVSVSEAGEG 588
>CO979463
Length = 851
Score = 72.8 bits (177), Expect = 2e-13
Identities = 36/95 (37%), Positives = 51/95 (52%), Gaps = 7/95 (7%)
Frame = -1
Query: 77 LDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDL 136
LD ++++ PTF +S+ + +CA+CL EF D R+L C H FH CID
Sbjct: 674 LDPSVIKSLPTFTFSAATHRSLQ-----DCAVCLSEFADGDEGRVLPNCKHAFHAHCIDT 510
Query: 137 WLESHKTCPVCRTDL-------DSPPNQMSKHGEG 164
W SH CP+CRT + D+ P +S+ GEG
Sbjct: 509 WFGSHSKCPLCRTPVLPATGSADTEPGSVSEAGEG 405
>TC208347 similar to UP|Q9XF63 (Q9XF63) RING-H2 zinc finger protein ATL3
(At1g72310) (RING-H2 zinc finger protein (ATL3);
86824-85850) (RING-H2 zinc finger protein ATL3;
90350-91324), partial (24%)
Length = 1229
Score = 70.9 bits (172), Expect = 7e-13
Identities = 45/131 (34%), Positives = 62/131 (46%), Gaps = 4/131 (3%)
Frame = +3
Query: 27 LTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVR----LSPDRSPSRGLDNTLL 82
+ ++I+ FV V+ +C + FA F W S S R S GLD +L
Sbjct: 264 VVIIIMFFVI--VIALCLHLFARNFW-WRSPAPQSRSHRRRRFVFSSGPDGGSGLDPAVL 434
Query: 83 EKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHK 142
P ++ K+ LECA+CL E + RLL C H FH CID+W +SH
Sbjct: 435 SSLPVLVFEGHAQEFKD---GLECAVCLSEVVEGEKARLLPKCNHGFHVACIDMWFQSHS 605
Query: 143 TCPVCRTDLDS 153
TCP+CR + S
Sbjct: 606 TCPLCRNPVAS 638
>TC232709 weakly similar to UP|Q9LZJ6 (Q9LZJ6) RING-H2 zinc finger protein
ATL5 (At3g62690), partial (19%)
Length = 987
Score = 70.5 bits (171), Expect = 9e-13
Identities = 49/172 (28%), Positives = 76/172 (43%), Gaps = 12/172 (6%)
Frame = +3
Query: 24 AFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQR--------TTSGSLVRLSPDRSPSR 75
AF+L +IL + F +C F +R +T + + +S +
Sbjct: 135 AFSLFGMILLIIIFH------FCVKYFIRRQQRRRQNDLLYQISTQIAPIDVSSVEPRNS 296
Query: 76 GLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQECID 135
GLD ++ P LY D K+ + C++CL +D++ R+L C H+FH +C+D
Sbjct: 297 GLDPLIIASLPKLLYKQT-DQFKQGEEVV*CSVCLGTIVEDTISRVLPNCKHIFHVDCVD 473
Query: 136 LWLESHKTCPVCRTDLDSPPNQMSKHGEGN----HNNNNNNLNVQEGMTSLP 183
W S+ TCP+CRT +D P +HG HN EG LP
Sbjct: 474 KWFNSNTTCPICRTVVD--PKVQPEHGHLGATRLHNQVQPTAPPAEGGDELP 623
>TC220950 similar to UP|Q9LS98 (Q9LS98) Similarity to RING zinc finger
protein, partial (57%)
Length = 719
Score = 68.2 bits (165), Expect(2) = 1e-12
Identities = 34/78 (43%), Positives = 42/78 (53%)
Frame = +2
Query: 71 RSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFH 130
R + G+ L+ FPT YS+ +L S EC ICL EF +R+L C H FH
Sbjct: 143 RVANTGVKKKALKTFPTVSYSAELNL---PSLDSECVICLSEFTSGDKVRILPKCNHRFH 313
Query: 131 QECIDLWLESHKTCPVCR 148
CID WL SH +CP CR
Sbjct: 314 VRCIDKWLSSHSSCPKCR 367
Score = 21.9 bits (45), Expect(2) = 1e-12
Identities = 7/8 (87%), Positives = 8/8 (99%)
Frame = +3
Query: 165 NHNNNNNN 172
+HNNNNNN
Sbjct: 426 HHNNNNNN 449
>TC212313
Length = 700
Score = 67.8 bits (164), Expect = 6e-12
Identities = 26/54 (48%), Positives = 35/54 (64%)
Frame = +2
Query: 105 ECAICLLEFDDDSMLRLLTICCHVFHQECIDLWLESHKTCPVCRTDLDSPPNQM 158
+C++CL EF +D LRLL C H FH CID WL SH CP+CR + + P ++
Sbjct: 17 DCSVCLSEFQEDESLRLLPKCNHAFHLPCIDTWLRSHTNCPMCRAPIVTDPTRV 178
>TC207001 similar to PIR|F86340|F86340 protein F2D10.34 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(54%)
Length = 1074
Score = 66.6 bits (161), Expect = 1e-11
Identities = 43/138 (31%), Positives = 61/138 (44%)
Frame = +2
Query: 11 PPAPTYKTPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPD 70
P A ++ ++ L ++C V V C W Q + GS R +
Sbjct: 242 PEAVALESDFVVILAALLCALICVVGLVAVARC---------AWFRQGSGGGSSPRQA-- 388
Query: 71 RSPSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFH 130
++GL +L+ P F Y V + + ECAICL +F +R+L C H FH
Sbjct: 389 -LANKGLKKKVLQSLPKFAY--VDSNPSKWVATSECAICLADFAAGDEIRVLPQCGHGFH 559
Query: 131 QECIDLWLESHKTCPVCR 148
CID WL SH +CP CR
Sbjct: 560 VPCIDTWLGSHSSCPSCR 613
>TC224907 weakly similar to UP|Q6Z8A6 (Q6Z8A6) Zinc finger (C3HC4-type RING
finger)-like, partial (12%)
Length = 1685
Score = 66.6 bits (161), Expect = 1e-11
Identities = 46/150 (30%), Positives = 64/150 (42%), Gaps = 9/150 (6%)
Frame = -1
Query: 11 PPAPTYKTPPALVAFTLTVLILCFVAFSVVYVCKYCFAGFFHTWALQRTTSGSLVRLSPD 70
PP P + PP+ V + +L +A + Y F F + +R
Sbjct: 1544 PPPPPFAGPPSSVDLSPLEFLLALLAVVTIPALIYTFIFAFGCPSRRRR----------- 1398
Query: 71 RSPSRGLDNTLLE--KFPTFLYSSVKDL----RKE---KSYSLECAICLLEFDDDSMLRL 121
R PS G + E F ++V D RKE K EC +CL F + +R
Sbjct: 1397 REPSYGEPSVASEVSHHQEFEIAAVADTEVKYRKEAHAKEIGGECPVCLSVFANGEEVRQ 1218
Query: 122 LTICCHVFHQECIDLWLESHKTCPVCRTDL 151
L+ C H FH CIDLWL +H CP+CR +
Sbjct: 1217 LSACKHSFHASCIDLWLSNHSNCPICRATI 1128
>TC228349
Length = 1112
Score = 66.6 bits (161), Expect = 1e-11
Identities = 48/140 (34%), Positives = 61/140 (43%), Gaps = 2/140 (1%)
Frame = +2
Query: 15 TYKTPPALVAFTLTVLILCFVAFSV--VYVCKYCFAGFFHTWALQRTTSGSLVRLSPDRS 72
T+K L + L + + C V VY+C +A H Q G V+ D+
Sbjct: 296 TFKGQKMLDSVLLALFLPCLGMSGVFIVYMCLLWYATTRHHQ--QPPIDGQPVKPVADK- 466
Query: 73 PSRGLDNTLLEKFPTFLYSSVKDLRKEKSYSLECAICLLEFDDDSMLRLLTICCHVFHQE 132
GL LEK P + KE ECA+CL E + + RL+ C H FH
Sbjct: 467 ---GLSALELEKLP-------RVTGKELVLGNECAVCLDEIESEQPARLVPGCNHGFHVH 616
Query: 133 CIDLWLESHKTCPVCRTDLD 152
C D WL H CPVCRT LD
Sbjct: 617 CADTWLSKHPLCPVCRTKLD 676
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,523,182
Number of Sequences: 63676
Number of extensions: 331145
Number of successful extensions: 3806
Number of sequences better than 10.0: 283
Number of HSP's better than 10.0 without gapping: 2916
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3469
length of query: 306
length of database: 12,639,632
effective HSP length: 97
effective length of query: 209
effective length of database: 6,463,060
effective search space: 1350779540
effective search space used: 1350779540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC126787.10


