

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.6 - phase: 0
(228 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
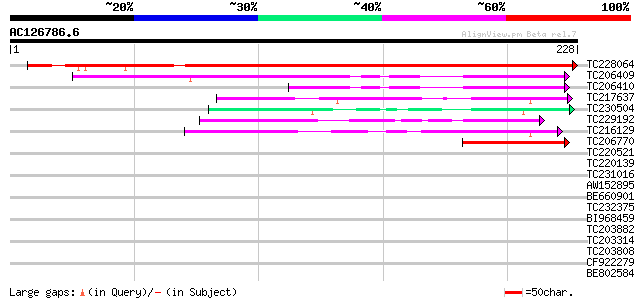
Score E
Sequences producing significant alignments: (bits) Value
TC228064 similar to UP|FKB1_ARATH (Q9LM71) Probable FKBP-type pe... 325 1e-89
TC206409 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type ... 102 1e-22
TC206410 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type ... 67 7e-12
TC217637 similar to GB|CAD35362.1|21535744|ATH490171 FK506 bindi... 48 4e-06
TC230504 similar to UP|Q9SCY3 (Q9SCY3) FKBP like protein , part... 44 5e-05
TC229192 similar to UP|Q6ZLL2 (Q6ZLL2) Immunophilin/FKBP-type pe... 44 6e-05
TC216129 44 6e-05
TC206770 similar to UP|Q93ZG9 (Q93ZG9) AT4g25340/T30C3_20, parti... 43 1e-04
TC220521 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl... 35 0.028
TC220139 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl... 35 0.028
TC231016 35 0.028
AW152895 33 0.11
BE660901 similar to GP|10177347|db peptidylprolyl isomerase {Ara... 32 0.18
TC232375 similar to UP|Q9FJL3 (Q9FJL3) Peptidylprolyl isomerase,... 32 0.18
BI968459 similar to GP|6686800|emb| FKBP like protein {Arabidops... 32 0.31
TC203882 similar to UP|FKB2_VICFA (Q41649) FK506-binding protein... 32 0.31
TC203314 homologue to UP|FKB2_VICFA (Q41649) FK506-binding prote... 31 0.53
TC203808 similar to UP|O22669 (O22669) Chlorophyll a/b binding p... 31 0.53
CF922279 29 1.5
BE802584 homologue to GP|20453145|gb| AT3g10060/T22K18_11 {Arabi... 27 5.9
>TC228064 similar to UP|FKB1_ARATH (Q9LM71) Probable FKBP-type
peptidyl-prolyl cis-trans isomerase 1, chloroplast
precursor (PPIase) (Rotamase) , partial (66%)
Length = 1043
Score = 325 bits (832), Expect = 1e-89
Identities = 168/232 (72%), Positives = 189/232 (81%), Gaps = 11/232 (4%)
Frame = +3
Query: 8 CSCSSSSLTWLLKKRNVDH-YGG------FSLRRRALPVVALSQS----RRSTAILISSL 56
CSC+ +S W +VD+ YGG +R R ++++ S RRSTA++ISSL
Sbjct: 75 CSCACASTNW-----SVDYGYGGGGVLSNSKVRSRRSKEISMAHSVCGSRRSTALVISSL 239
Query: 57 PFTFVFLSPPPPAEARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFD 116
PF F+FLSPP A RR KK IP D YITSPDGLKYYD +EGKGP+AEKG+TVQVHFD
Sbjct: 240 PFGFLFLSPP----AEARRNKKAIPEDQYITSPDGLKYYDLVEGKGPVAEKGTTVQVHFD 407
Query: 117 CLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPP 176
CLYRGITAVSSRESKLLAGNR+IAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPP
Sbjct: 408 CLYRGITAVSSRESKLLAGNRIIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPP 587
Query: 177 QAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQLAST 228
A+Y IVEGMRVGGKRTVIVPPE GYG+KGMNEIPPGATFELN+ELLQ+ +T
Sbjct: 588 PAVYIIVEGMRVGGKRTVIVPPENGYGQKGMNEIPPGATFELNVELLQVVAT 743
>TC206409 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type
peptidyl-prolyl cis-trans isomerase-like, partial (57%)
Length = 983
Score = 102 bits (254), Expect = 1e-22
Identities = 75/204 (36%), Positives = 97/204 (46%), Gaps = 4/204 (1%)
Frame = +1
Query: 26 HYGGFSLRRRALPVVALSQSRRSTAILISSLPFTFVFLSPPPPAEA----RERRKKKNIP 81
H S + A V +S S L+S L T V + A A R + IP
Sbjct: 169 HCSHSSTNKIAAEPVTVSLSIEGRRALLSCLLTTVVGVYACDVAGAVSTSRRALRGAKIP 348
Query: 82 IDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGNRVIAQ 141
DY T P+GLKYYD G G A+KGS V +H+ ++ IT ++SR+ + G
Sbjct: 349 ESDYTTLPNGLKYYDLKVGNGAEAKKGSRVAIHYVAKWKSITFMTSRQGMGVGGGT---- 516
Query: 142 PYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKG 201
PY F VG ER G V+GMRVGG+R +IVPPE
Sbjct: 517 PYGFDVGQ---SERGTVLKGLDLG-----------------VQGMRVGGQRLLIVPPELA 636
Query: 202 YGKKGMNEIPPGATFELNIELLQL 225
YG KG+ EIPP +T EL+IELL +
Sbjct: 637 YGSKGVQEIPPNSTIELDIELLSI 708
>TC206410 similar to UP|Q6Z4E9 (Q6Z4E9) Immunophilin / FKBP-type
peptidyl-prolyl cis-trans isomerase-like, partial (36%)
Length = 539
Score = 67.0 bits (162), Expect = 7e-12
Identities = 43/113 (38%), Positives = 58/113 (51%)
Frame = +2
Query: 113 VHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAA 172
+H+ ++GIT ++SR+ + G PY F VG ER G
Sbjct: 83 IHYVAKWKGITFMTSRQGMGVGGGT----PYGFDVGQ---SERGTVLKGLDLG------- 220
Query: 173 PKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
V+GMRVGG+R +IVPPE YG KG+ EIPP +T EL+IELL +
Sbjct: 221 ----------VQGMRVGGQRLLIVPPELAYGSKGVQEIPPNSTIELDIELLSI 349
>TC217637 similar to GB|CAD35362.1|21535744|ATH490171 FK506 binding protein 1
{Arabidopsis thaliana;} , partial (60%)
Length = 870
Score = 47.8 bits (112), Expect = 4e-06
Identities = 41/150 (27%), Positives = 64/150 (42%), Gaps = 7/150 (4%)
Frame = +1
Query: 84 DYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRES-KLLAGNRVIAQP 142
++ +P GL + D L G GP A KG ++ H+ V E+ K+ + +P
Sbjct: 289 EFQVAPSGLAFCDKLVGAGPQAVKGQLIKAHY---------VGRLENGKVFDSSYNRGKP 441
Query: 143 YEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGY 202
F+VG + E + +G+ PP M GGKRT+ +PPE GY
Sbjct: 442 LTFRVGVGEVIKGWDEGIIGGDGV--------PP---------MLAGGKRTLKIPPELGY 570
Query: 203 GKKGMN------EIPPGATFELNIELLQLA 226
G +G IPP + ++E + A
Sbjct: 571 GSRGAGCRGGSCIIPPDSVLLFDVEFVSKA 660
>TC230504 similar to UP|Q9SCY3 (Q9SCY3) FKBP like protein , partial (68%)
Length = 988
Score = 44.3 bits (103), Expect = 5e-05
Identities = 43/154 (27%), Positives = 62/154 (39%), Gaps = 7/154 (4%)
Frame = +1
Query: 81 PIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYR-GITAVSSRESKLLAGNRVI 139
P+ +Y GL Y D EG G A G + VH+ + GI SS +
Sbjct: 238 PLCEYNYVKSGLGYCDIAEGFGDEAPLGELINVHYTARFADGIVFDSSYKR--------- 390
Query: 140 AQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPE 199
A+P ++G GK K G+ A+ P MR+GGKR + +PP
Sbjct: 391 ARPLTMRIGV--GKVIK----GLDQGILGAEGXPP-----------MRIGGKRQLQIPPH 519
Query: 200 KGYGKK------GMNEIPPGATFELNIELLQLAS 227
YG + G IP AT +I +++ S
Sbjct: 520 LAYGPEPAGCFSGDCNIPANATLLYDINFVEVYS 621
>TC229192 similar to UP|Q6ZLL2 (Q6ZLL2) Immunophilin/FKBP-type
peptidyl-prolyl cis-trans isomerase-like protein,
partial (65%)
Length = 1206
Score = 43.9 bits (102), Expect = 6e-05
Identities = 40/139 (28%), Positives = 57/139 (40%)
Frame = +1
Query: 77 KKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRESKLLAGN 136
K + DY + GL+Y D G GP + G TV V +D G
Sbjct: 358 KTKMRYPDYTETESGLQYKDLRPGNGPKPKMGETVVVDWDGYTIGYYG------------ 501
Query: 137 RVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIV 196
R+ + K G+ G ++ +F G + P +A V GM +GG R +IV
Sbjct: 502 RIFEARNKTKGGSFEGDDK--DFFKFKIGY--NEVIPAFEEA----VSGMALGGIRRIIV 657
Query: 197 PPEKGYGKKGMNEIPPGAT 215
PPE GY + N+ P T
Sbjct: 658 PPELGYPENDFNKSGPRPT 714
>TC216129
Length = 1001
Score = 43.9 bits (102), Expect = 6e-05
Identities = 35/159 (22%), Positives = 67/159 (42%), Gaps = 7/159 (4%)
Frame = +3
Query: 71 ARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHFDCLYRGITAVSSRES 130
+++ +N+ + + P+G++YY+ G G G V +
Sbjct: 444 SQQEANTRNVEKVEEVVLPNGIRYYELKVGGGASPRPGDLVVIDIT-------------G 584
Query: 131 KLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGG 190
K+ + V +E G ++ V + S + + + +++ M+ GG
Sbjct: 585 KIESSGEVFVNTFE-------GDKKPLALV-----MGSRPYSKGVCEGIEYVLKTMKAGG 728
Query: 191 KRTVIVPPEKGYGKKGMN-------EIPPGATFELNIEL 222
KR VIVPP+ G+G+ G + +IPP AT E +E+
Sbjct: 729 KRKVIVPPQLGFGENGADFDSGTGVQIPPLATLEYILEV 845
>TC206770 similar to UP|Q93ZG9 (Q93ZG9) AT4g25340/T30C3_20, partial (23%)
Length = 929
Score = 43.1 bits (100), Expect = 1e-04
Identities = 16/43 (37%), Positives = 26/43 (60%)
Frame = +2
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGATFELNIELLQL 225
+ GMR+G KR + +PP GY K + IPP + ++EL+ +
Sbjct: 536 INGMRIGDKRRITIPPSMGYADKRVGSIPPNSWLVFDVELVDV 664
>TC220521 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl isomerase
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) , partial (40%)
Length = 890
Score = 35.0 bits (79), Expect = 0.028
Identities = 17/46 (36%), Positives = 25/46 (53%), Gaps = 1/46 (2%)
Frame = +1
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNE-IPPGATFELNIELLQLAS 227
++ M+ G +PPE YG+ G IPP AT + ++ELL S
Sbjct: 448 IKTMKKGENALFTIPPELAYGESGSPPTIPPNATLQFDVELLSWTS 585
>TC220139 similar to UP|FKB7_WHEAT (Q43207) 70 kDa peptidylprolyl isomerase
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) , partial (29%)
Length = 722
Score = 35.0 bits (79), Expect = 0.028
Identities = 17/46 (36%), Positives = 25/46 (53%), Gaps = 1/46 (2%)
Frame = +1
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNE-IPPGATFELNIELLQLAS 227
++ M+ G +PPE YG+ G IPP AT + ++ELL S
Sbjct: 376 IKTMKKGENAIFTIPPELAYGESGSPPTIPPNATLQFDVELLSWTS 513
>TC231016
Length = 851
Score = 35.0 bits (79), Expect = 0.028
Identities = 18/48 (37%), Positives = 25/48 (51%)
Frame = +2
Query: 68 PAEARERRKKKNIPIDDYITSPDGLKYYDFLEGKGPIAEKGSTVQVHF 115
P RE + K + D+YI S GL Y DF+ G+G + G V H+
Sbjct: 65 PNFVREGFQVKVVSSDNYIKSDSGLIYRDFVVGQGDCPKDGQQVTFHY 208
Score = 29.3 bits (64), Expect = 1.5
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = +3
Query: 183 VEGMRVGGKRTVIVPPEKG 201
+ MR GGKR +I+PPE G
Sbjct: 318 IRDMRPGGKRRIIIPPELG 374
>AW152895
Length = 411
Score = 33.1 bits (74), Expect = 0.11
Identities = 16/38 (42%), Positives = 21/38 (55%)
Frame = +2
Query: 178 AMYTIVEGMRVGGKRTVIVPPEKGYGKKGMNEIPPGAT 215
A+ V M +GG +T+IVPPE GY N+ P T
Sbjct: 107 ALEDAVSDMALGGIKTIIVPPELGYPDNDCNKSGPRPT 220
>BE660901 similar to GP|10177347|db peptidylprolyl isomerase {Arabidopsis
thaliana}, partial (34%)
Length = 724
Score = 32.3 bits (72), Expect = 0.18
Identities = 39/141 (27%), Positives = 52/141 (36%), Gaps = 3/141 (2%)
Frame = +2
Query: 90 DGLKYYDFLEGKG-PIAEKGSTVQVHFD-CLYRGITAVSSRESKLLAGNRVIAQPYEFKV 147
+GLK EG+G E G VQVH+ L G SSR+ P+ F +
Sbjct: 116 EGLKKKLLKEGEGWDTPEVGDEVQVHYTGTLLDGTKFDSSRDR---------GTPFSFTL 268
Query: 148 GAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM 207
G Q Q + T M+ G +P E YG+ G
Sbjct: 269 G-------------------QGQVIKGWDQGIIT----MKKGENSLFTIPAELAYGETGS 379
Query: 208 NE-IPPGATFELNIELLQLAS 227
IPP AT + ++ELL S
Sbjct: 380 PPTIPPNATLQFDVELLSWTS 442
>TC232375 similar to UP|Q9FJL3 (Q9FJL3) Peptidylprolyl isomerase, partial
(21%)
Length = 559
Score = 32.3 bits (72), Expect = 0.18
Identities = 38/137 (27%), Positives = 51/137 (36%), Gaps = 3/137 (2%)
Frame = +3
Query: 90 DGLKYYDFLEGKG-PIAEKGSTVQVHFD-CLYRGITAVSSRESKLLAGNRVIAQPYEFKV 147
+GLK EG+G E G VQVH+ L G SSR+ P+ F +
Sbjct: 234 EGLKKKLLKEGEGWDTPEAGDEVQVHYTGTLLDGTKFDSSRDR---------GTPFSFTL 386
Query: 148 GAPPGKERKREFVDNPNGLFSAQAAPKPPQAMYTIVEGMRVGGKRTVIVPPEKGYGKKGM 207
G Q Q + T M+ G +P E YG+ G
Sbjct: 387 G-------------------QGQVIKGWDQGIIT----MKKGENALFTIPAELAYGESGS 497
Query: 208 NE-IPPGATFELNIELL 223
IPP AT + ++ELL
Sbjct: 498 PPTIPPNATLQFDVELL 548
>BI968459 similar to GP|6686800|emb| FKBP like protein {Arabidopsis
thaliana}, partial (17%)
Length = 591
Score = 31.6 bits (70), Expect = 0.31
Identities = 14/30 (46%), Positives = 16/30 (52%)
Frame = -3
Query: 183 VEGMRVGGKRTVIVPPEKGYGKKGMNEIPP 212
V M+VGG R VI+PP Y IPP
Sbjct: 412 VRSMKVGGIRRVIIPPSLEYQNTSQEPIPP 323
>TC203882 similar to UP|FKB2_VICFA (Q41649) FK506-binding protein 2 precursor
(Peptidyl-prolyl cis-trans isomerase) (PPIase)
(Rotamase) (15 kDa FKBP) (FKBP-15) , partial (85%)
Length = 944
Score = 31.6 bits (70), Expect = 0.31
Identities = 17/40 (42%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Frame = +2
Query: 185 GMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELL 223
GM +G KR + +P + GYG++G IP GAT + EL+
Sbjct: 398 GMCLGEKRKLKIPSKLGYGEQGSPPTIPGGATLIFDAELV 517
>TC203314 homologue to UP|FKB2_VICFA (Q41649) FK506-binding protein 2
precursor (Peptidyl-prolyl cis-trans isomerase)
(PPIase) (Rotamase) (15 kDa FKBP) (FKBP-15) , partial
(85%)
Length = 746
Score = 30.8 bits (68), Expect = 0.53
Identities = 17/40 (42%), Positives = 24/40 (59%), Gaps = 1/40 (2%)
Frame = +2
Query: 185 GMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIELL 223
GM +G KR + +P + GYG +G IP GAT + EL+
Sbjct: 335 GMCLGEKRKLKIPAKLGYGDQGSPPTIPGGATLIFDTELV 454
>TC203808 similar to UP|O22669 (O22669) Chlorophyll a/b binding protein of
LHCII type I precursor, partial (25%)
Length = 586
Score = 30.8 bits (68), Expect = 0.53
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Frame = -3
Query: 185 GMRVGGKRTVIVPPEKGYGKKGM-NEIPPGATFELNIEL 222
GM +G KR + +P + GYG +G IP GAT + EL
Sbjct: 236 GMGLGEKRKLKIPAKLGYGDQGSPPTIPGGATLIFDTEL 120
>CF922279
Length = 528
Score = 29.3 bits (64), Expect = 1.5
Identities = 22/69 (31%), Positives = 33/69 (46%), Gaps = 2/69 (2%)
Frame = +3
Query: 146 KVGAPPGKERKREFVDNPNGLFSAQAA-PKPPQAMY-TIVEGMRVGGKRTVIVPPEKGYG 203
K G PP ++ K F P G Q + +PP+ ++ T G + GG PP+K G
Sbjct: 153 KGGGPPPEKTKNFFKTPPPGAPRGQKSFYRPPKRVFQTPPLGAQPGG------PPQKKKG 314
Query: 204 KKGMNEIPP 212
+K + PP
Sbjct: 315 EKNPPQFPP 341
>BE802584 homologue to GP|20453145|gb| AT3g10060/T22K18_11 {Arabidopsis
thaliana}, partial (23%)
Length = 324
Score = 27.3 bits (59), Expect = 5.9
Identities = 24/81 (29%), Positives = 34/81 (41%)
Frame = +2
Query: 112 QVHFDCLYRGITAVSSRESKLLAGNRVIAQPYEFKVGAPPGKERKREFVDNPNGLFSAQA 171
Q+H+ ++GIT ++SR+ + G PY F VG ER G
Sbjct: 152 QIHYVAKWKGITFMTSRQGMGVGG----GTPYGFDVGQ---SERGTVLKGLDLG------ 292
Query: 172 APKPPQAMYTIVEGMRVGGKR 192
V+GMRVGG+R
Sbjct: 293 -----------VQGMRVGGQR 322
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,224,379
Number of Sequences: 63676
Number of extensions: 184210
Number of successful extensions: 1450
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 1340
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1435
length of query: 228
length of database: 12,639,632
effective HSP length: 94
effective length of query: 134
effective length of database: 6,654,088
effective search space: 891647792
effective search space used: 891647792
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC126786.6


