

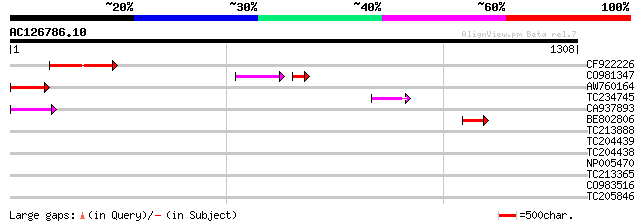
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126786.10 - phase: 0 /pseudo
(1308 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922226 123 5e-28
CO981347 97 6e-25
AW760164 similar to GP|11994422|dbj oxidoreductase short-chain ... 97 4e-20
TC234745 weakly similar to UP|Q9FFM0 (Q9FFM0) Copia-like retrotr... 66 9e-11
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 54 5e-07
BE802806 45 2e-04
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 37 0.044
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 34 0.48
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 34 0.48
NP005470 reverse transcriptase 32 1.8
TC213365 similar to UP|Q6I8L6 (Q6I8L6) Reverse transcriptase (Fr... 30 9.1
CO983516 30 9.1
TC205846 homologue to UP|Q6X7J9 (Q6X7J9) WOX4 protein (At1g46480... 28 9.8
>CF922226
Length = 667
Score = 123 bits (309), Expect = 5e-28
Identities = 64/155 (41%), Positives = 101/155 (64%)
Frame = -3
Query: 93 MTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDNLANIDVNLEDEDKALHLLCAL 152
MTKSL +R KQ LY ++M E + + EQL FNK+I +L NIDV ++DED+AL LLC L
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 153 PRSFENFKDTMLYGKEGTITLEEVQAALRTKELTKFKELKVDDSGEGLNVSRGRSQNRGK 212
P+S+ +FK+T+L+G++ +++L+EVQ AL +KEL + KE K SGEGL + +
Sbjct: 485 PKSYSHFKETLLFGRD-SVSLDEVQTALNSKELNERKEKKSSASGEGLTARGKTFKKDSE 309
Query: 213 GKGKNSRSKSRSKGDGNKTQYKCFICHNLVTSRRI 247
K + +++ G+GN + +C+ C +R++
Sbjct: 308 FDKKKQKPENQKNGEGNIFKIRCYHCKKEGHTRKV 204
>CO981347
Length = 624
Score = 97.4 bits (241), Expect(2) = 6e-25
Identities = 56/113 (49%), Positives = 68/113 (59%)
Frame = +1
Query: 522 KIQRMGYTCRKSDWN*TESVEN*QWPEVCFRAV**VLQEERYKEA*NRGIHTSTEWSC*K 581
KIQRM Y+ KS W +S ++*QWP VCFRAV *VLQE R+++A N HT EW K
Sbjct: 112 KIQRMTYSYWKSTWYKIKSFKD*QWPGVCFRAVQ*VLQENRHQKAQNSPSHTIAEWFSRK 291
Query: 582 NEQDFVGACEVYAAGSWIVQEFLGRGC*YCSIFD*QMSINRDRSQDTYGGLEW 634
+E D G EV+A ++ LGR C + IFD* M R QDT G LEW
Sbjct: 292 DEYDHFGKSEVHATKCKTAKDLLGRSCKHNIIFD**MPFINLRFQDTNGSLEW 450
Score = 36.6 bits (83), Expect(2) = 6e-25
Identities = 18/40 (45%), Positives = 26/40 (65%)
Frame = +2
Query: 652 SCQTRQT*C*SCEVYFHGLS*RCERLQTVEDGTWRIKIYY 691
SC TR+ C C+V H LS*R ++Q +E GTW K+++
Sbjct: 503 SC*TRKAGCKGCKVCVHWLS*RS*KVQAMEIGTW*DKMHH 622
>AW760164 similar to GP|11994422|dbj oxidoreductase short-chain
dehydrogenase/reductase family-like protein {Arabidopsis
thaliana}, partial (9%)
Length = 428
Score = 97.4 bits (241), Expect = 4e-20
Identities = 52/91 (57%), Positives = 68/91 (74%), Gaps = 1/91 (1%)
Frame = +3
Query: 2 MGS-K*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAV 60
MGS K ++EKFTG NDFGL +KMRA+L+QQ VEAL GE ++ + +K L KA
Sbjct: 75 MGSAKYEVEKFTGQNDFGLC*LKMRALLVQQGLVEALDGEIKLEKMMADGDKKALLQKAY 254
Query: 61 SAIIMCLGDKVLREVSRETTAVSMWNKLDSL 91
+AII+ LGDKVLR+VS+ETTAV +W+KL+ L
Sbjct: 255 NAIILSLGDKVLRQVSKETTAVGVWSKLEVL 347
>TC234745 weakly similar to UP|Q9FFM0 (Q9FFM0) Copia-like retrotransposable
element, partial (5%)
Length = 494
Score = 66.2 bits (160), Expect = 9e-11
Identities = 40/90 (44%), Positives = 51/90 (56%)
Frame = -2
Query: 835 SGKQVGVQEKRRYTGGRSTKV*GKTCSKGFYSGGRD*LQ*NLFTGGETLFYKGTYGDC*H 894
S QVG+QEK +++ +V GKTC GFY+ G+ *L *N T G+TLFYK G+C
Sbjct: 265 SWMQVGLQEKEKHSKCEEVEVQGKTCG*GFYTSGQS*L**NFLTCGKTLFYKDLDGNCKS 86
Query: 895 V*S*VRTDGCEDRISTWRVGRNYLYATTRR 924
V*S V GC+D N+ Y T R
Sbjct: 85 V*SGVTIIGCQD---------NFFYMETLR 23
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 53.9 bits (128), Expect = 5e-07
Identities = 32/106 (30%), Positives = 55/106 (51%)
Frame = -2
Query: 3 GSK*DIEKFTGGNDFGLWKVKMRAILIQQKCVEALKGEAQMAAHLTPAEKTELNDKAVSA 62
G+K ++ KF G +F LW+ +++ +L Q ++ L+ E EL ++ +
Sbjct: 315 GAKFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRDSKSNNTEALDWE--EL*ERTATT 142
Query: 63 IIMCLGDKVLREVSRETTAVSMWNKLDSLYMTKSLAHRQCLKQQLY 108
I +CL D+ L + +W KL+S YM KSL ++ L Q+LY
Sbjct: 141 IRLCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>BE802806
Length = 285
Score = 45.4 bits (106), Expect = 2e-04
Identities = 23/58 (39%), Positives = 36/58 (61%)
Frame = -1
Query: 1046 EESGGAVQNERF*SR*HSSWSPHKAINKAVSSIR*REKQDGRYSLCKWGWKHHVWYGL 1103
+ESGG V + + *H++W +KAI + + *R+ + +LC+W WKH+VW GL
Sbjct: 189 QESGGKV*DASKQTC*HTTWPSYKAICHSSTRNS*RKV*NESNTLCQWCWKHNVWNGL 16
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 37.4 bits (85), Expect = 0.044
Identities = 19/47 (40%), Positives = 26/47 (54%)
Frame = +2
Query: 1245 FGKSSGIS*KDKTH*HSFALRQRHD*DKGDHDRESGIGRQSSRHVHQ 1291
FGK S +S*K TH H L +R *++G + + R S RH H+
Sbjct: 236 FGKKSSLS*KK*THRHPLPLHKRMH*EEGGKVKVCDVSRSSCRHFHK 376
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 33.9 bits (76), Expect = 0.48
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Frame = +3
Query: 902 DGCEDRISTWRVGRNYLYATTRRFCRRQYKSMFV--EEIFVWVEAKSKAVV 950
DGCE+RIS W L + CR + +E +W+EA SK++V
Sbjct: 3522 DGCEERISEWIPE*RSLCGAAKGICRPDSSRSCIQAQEGSLWIEASSKSLV 3674
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 33.9 bits (76), Expect = 0.48
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Frame = +3
Query: 902 DGCEDRISTWRVGRNYLYATTRRFCRRQYKSMFV--EEIFVWVEAKSKAVV 950
DGCE+R+S W L + CR + +E +W+EA SK++V
Sbjct: 3525 DGCEERVSEWIPE*RSLCGAAKGICRSNSSRSCIQAQEGSLWIEASSKSLV 3677
>NP005470 reverse transcriptase
Length = 267
Score = 32.0 bits (71), Expect = 1.8
Identities = 28/88 (31%), Positives = 40/88 (44%), Gaps = 2/88 (2%)
Frame = +3
Query: 906 DRISTWRVGRNYLYATTRRF--CRRQYKSMFVEEIFVWVEAKSKAVVSSV**VPSKGWFC 963
D + ++GR Y T R R++ E+ VW E SKA+V * + +
Sbjct: 3 DSVFARKIGRKYSNETA*RV*SSRKRKVCFSTAEVLVWFETISKAMVHEF*QLYHQPRIQ 182
Query: 964 EKQL**LCLHDEKE*ESHSLSSPLCR*Y 991
EK L* LCL + + LSS + *Y
Sbjct: 183 EKSL*LLCLSQQGGGWINDLSSAVRG*Y 266
>TC213365 similar to UP|Q6I8L6 (Q6I8L6) Reverse transcriptase (Fragment),
partial (10%)
Length = 440
Score = 29.6 bits (65), Expect = 9.1
Identities = 20/55 (36%), Positives = 29/55 (52%)
Frame = +2
Query: 1199 PFNNSSRVHSPC*RGQGSHMVERYDWRNGN*SRVCEDTL**PKCHSFGKSSGIS* 1253
P N ++ * +G ++ ER D + N*SR+C L* P C+ + SG S*
Sbjct: 14 PLNY*G*IYGSY*SCKGRNLAERSDK*SQN*SRIC*HLL*QP*CYMLRQGSGSS* 178
>CO983516
Length = 724
Score = 29.6 bits (65), Expect = 9.1
Identities = 16/51 (31%), Positives = 25/51 (48%), Gaps = 2/51 (3%)
Frame = +1
Query: 902 DGCEDRISTWRVGRNYLYATTRRFCRRQYKSMFV--EEIFVWVEAKSKAVV 950
DGCE+R+S W L + R + +E +W+EA SK++V
Sbjct: 442 DGCEERVSEWIPE*RSLCGAAKGIYRSNSSRSCIQAQEGSLWIEASSKSLV 594
>TC205846 homologue to UP|Q6X7J9 (Q6X7J9) WOX4 protein (At1g46480), partial
(41%)
Length = 877
Score = 28.1 bits (61), Expect(2) = 9.8
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = +1
Query: 223 RSKGDGNKTQYKCFICHNLVTSRRI 247
RS GDG T +K CH+LV R I
Sbjct: 562 RSAGDGYLTAWKNKTCHHLVNKRNI 636
Score = 19.6 bits (39), Expect(2) = 9.8
Identities = 10/22 (45%), Positives = 15/22 (67%)
Frame = +1
Query: 203 SRGRSQNRGKGKGKNSRSKSRS 224
S+GR+ + G K++R KSRS
Sbjct: 376 SKGRTCSIGFRITKHARDKSRS 441
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.360 0.161 0.601
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,343,461
Number of Sequences: 63676
Number of extensions: 847091
Number of successful extensions: 8138
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 4463
Number of HSP's successfully gapped in prelim test: 241
Number of HSP's that attempted gapping in prelim test: 3419
Number of HSP's gapped (non-prelim): 5059
length of query: 1308
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1200
effective length of database: 5,762,624
effective search space: 6915148800
effective search space used: 6915148800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC126786.10


