

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126782.6 - phase: 1
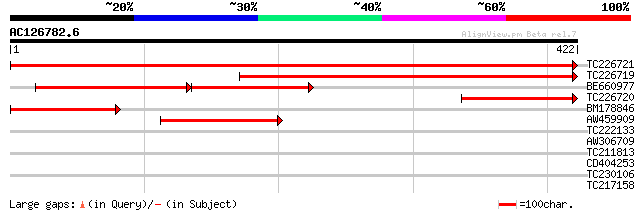
(422 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226721 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate syn... 734 0.0
TC226719 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate syn... 432 e-121
BE660977 211 2e-82
TC226720 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate syn... 147 7e-36
BM178846 126 2e-29
AW459909 115 4e-26
TC222133 30 2.7
AW306709 weakly similar to SP|P93329|NO20_ Early nodulin 20 prec... 29 3.5
TC211813 similar to UP|Q9FH76 (Q9FH76) Cytochrome P450 monooxyge... 29 4.6
CD404253 28 5.9
TC230106 homologue to UP|O80412 (O80412) Mitochondrial phosphate... 28 5.9
TC217158 similar to UP|Q9SUW2 (Q9SUW2) Adenine phosphoribosyltra... 28 5.9
>TC226721 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate synthase,
chloroplast precursor (Citrulline--aspartate ligase) ,
partial (76%)
Length = 1737
Score = 734 bits (1895), Expect = 0.0
Identities = 349/423 (82%), Positives = 398/423 (93%), Gaps = 1/423 (0%)
Frame = +3
Query: 1 IKAVLYKDVEI-SETKKVAGLKGKLNKVVLAYSGGLDTSVVVPWLRENYGCDVVCFTADL 59
IKA D E+ SET K GL+GKLNKVVLAYSGGLDTSV+VPWLRENYGC+VVCFTAD+
Sbjct: 228 IKAAARSDTEVVSETTKAGGLRGKLNKVVLAYSGGLDTSVIVPWLRENYGCEVVCFTADV 407
Query: 60 GQGASELEGLEEKAKASGASQLVVKDLKEEFVRDYVFPCLRAGAIYERKYLLGTAIARPV 119
GQG ELEGLE+KAKASGA QLVVKDLKEEFV+DY+FPCLRAGA+YERKYLLGT++ARPV
Sbjct: 408 GQGIKELEGLEQKAKASGACQLVVKDLKEEFVKDYIFPCLRAGAVYERKYLLGTSMARPV 587
Query: 120 IAKAMVDIANEVGADAVSHGCTGKGNDQVRFELSFFALNPKLNVVAPWREWDITGREDAI 179
IAKAMVD+A EVGADAVSHGCTGKGNDQVRFEL+FFALNPKLNVVAPWREWDITGREDAI
Sbjct: 588 IAKAMVDVAKEVGADAVSHGCTGKGNDQVRFELTFFALNPKLNVVAPWREWDITGREDAI 767
Query: 180 EYAKKHDVPVPVTKKSIYSRDRNLWHITHEGDVLEDPANEPKKDMYMLTVDPEDAPNEPE 239
EYAKKH++PVPVTK SIYSRDRNLWH++HEGD+LEDPANEPKKDMY+++VDPEDAP++PE
Sbjct: 768 EYAKKHNIPVPVTKTSIYSRDRNLWHLSHEGDILEDPANEPKKDMYIMSVDPEDAPDQPE 947
Query: 240 YVEIGFELGIPVSLNGKRLSPGNLVAELNEIGGRHGIGRVDLVEDRIVGMKSRGVYETPG 299
YVEIG E G+PVS+NG+RLSP +L+AELNEIGG+HGIGRVD+VE+R+VGMKSRGVYETPG
Sbjct: 948 YVEIGIESGLPVSVNGRRLSPASLLAELNEIGGKHGIGRVDMVENRLVGMKSRGVYETPG 1127
Query: 300 GTILFTAARDLETLTLDRETLQLKDSLALKYAELVYAGRWFDPLRESMDSFMEKISQTTT 359
GTILF AAR+LE+LTLDRET+Q+KDSLALKYAELVYAGRWFDPLRESMD+FM+KI++TTT
Sbjct: 1128GTILFAAARELESLTLDRETIQVKDSLALKYAELVYAGRWFDPLRESMDAFMQKITETTT 1307
Query: 360 GSVSLKLYKGSASVTGRKSPYSLYREDISSFESGEIYNHADAVGFIKLYGLPMRIRAMME 419
GSV+LKLYKGS + T R SP+SLYR+DISSFESG+IY+ ADA GFI+LYGLPMR+RAM+E
Sbjct: 1308GSVTLKLYKGSVTATSRTSPFSLYRQDISSFESGQIYDQADAAGFIRLYGLPMRVRAMLE 1487
Query: 420 QGI 422
GI
Sbjct: 1488HGI 1496
>TC226719 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate synthase,
chloroplast precursor (Citrulline--aspartate ligase) ,
partial (47%)
Length = 917
Score = 432 bits (1112), Expect = e-121
Identities = 205/251 (81%), Positives = 236/251 (93%)
Frame = +1
Query: 172 ITGREDAIEYAKKHDVPVPVTKKSIYSRDRNLWHITHEGDVLEDPANEPKKDMYMLTVDP 231
ITGREDAIEYAKKH++PVPVTKKSIYSRDRNLWH++HEGD+LEDPANEPKKDMYM++VD
Sbjct: 1 ITGREDAIEYAKKHNIPVPVTKKSIYSRDRNLWHLSHEGDILEDPANEPKKDMYMMSVDA 180
Query: 232 EDAPNEPEYVEIGFELGIPVSLNGKRLSPGNLVAELNEIGGRHGIGRVDLVEDRIVGMKS 291
EDAP+E EYVEI E G+PVS+NGKRLSP L+ ELNEIGGRHGIGR+D+VE+R+VGMKS
Sbjct: 181 EDAPDEAEYVEIEIESGLPVSVNGKRLSPATLLTELNEIGGRHGIGRIDMVENRLVGMKS 360
Query: 292 RGVYETPGGTILFTAARDLETLTLDRETLQLKDSLALKYAELVYAGRWFDPLRESMDSFM 351
GVYETPGGTILF AAR+LE LTLDRET+Q+KDSLALKYAELVYAGRWFDPLRESMD+FM
Sbjct: 361 PGVYETPGGTILFAAARELEFLTLDRETIQVKDSLALKYAELVYAGRWFDPLRESMDAFM 540
Query: 352 EKISQTTTGSVSLKLYKGSASVTGRKSPYSLYREDISSFESGEIYNHADAVGFIKLYGLP 411
+KI++TTTGSV+LKLYKGS +VT RKSP+SLYR+DISSFESG+IY+ ADA GFI+LYGLP
Sbjct: 541 QKITETTTGSVTLKLYKGSVTVTSRKSPFSLYRQDISSFESGQIYDQADAAGFIRLYGLP 720
Query: 412 MRIRAMMEQGI 422
MR+RAM+EQGI
Sbjct: 721 MRVRAMLEQGI 753
>BE660977
Length = 718
Score = 211 bits (536), Expect(2) = 2e-82
Identities = 102/116 (87%), Positives = 111/116 (94%)
Frame = +3
Query: 20 LKGKLNKVVLAYSGGLDTSVVVPWLRENYGCDVVCFTADLGQGASELEGLEEKAKASGAS 79
L+GKL+KVVLAYSGGLDTSV+VPWLRENYGC+VVCFTAD+GQG ELEGLE KAKASGAS
Sbjct: 72 LRGKLSKVVLAYSGGLDTSVIVPWLRENYGCEVVCFTADVGQGIQELEGLEAKAKASGAS 251
Query: 80 QLVVKDLKEEFVRDYVFPCLRAGAIYERKYLLGTAIARPVIAKAMVDIANEVGADA 135
QLVVKDLKEEFV DYVFPCLRAGA+YERKYLLGT++ARPVIAKAMVD+A EVGADA
Sbjct: 252 QLVVKDLKEEFVSDYVFPCLRAGAVYERKYLLGTSMARPVIAKAMVDVAKEVGADA 419
Score = 114 bits (284), Expect(2) = 2e-82
Identities = 62/94 (65%), Positives = 68/94 (71%), Gaps = 3/94 (3%)
Frame = +1
Query: 136 VSHGCTGKGNDQVRFELSFFALNPKLNVVAPWREWDITGREDAIEYAKKHDVPVPVTKKS 195
VSHGCTGKGNDQVRFEL+FFALNPKLNVVAP REWDITGREDAIEYAKKH K
Sbjct: 421 VSHGCTGKGNDQVRFELTFFALNPKLNVVAPXREWDITGREDAIEYAKKHKYSCSCDKDX 600
Query: 196 -IYSRDRNLW-HITHEGDVL-EDPANEPKKDMYM 226
I + D + + H G + PA EPKKDMY+
Sbjct: 601 HILAXD*GXFCXLXH*GRXFWKTPAYEPKKDMYI 702
>TC226720 similar to UP|ASSY_ARATH (Q9SZX3) Argininosuccinate synthase,
chloroplast precursor (Citrulline--aspartate ligase) ,
partial (16%)
Length = 406
Score = 147 bits (372), Expect = 7e-36
Identities = 68/86 (79%), Positives = 81/86 (94%)
Frame = +1
Query: 337 GRWFDPLRESMDSFMEKISQTTTGSVSLKLYKGSASVTGRKSPYSLYREDISSFESGEIY 396
GRWFDPLRESMD+FM+KI++TTTGSV+LKLYKGS +VT R SP+SLYREDISSFESG+IY
Sbjct: 1 GRWFDPLRESMDAFMQKITETTTGSVTLKLYKGSVTVTSRTSPFSLYREDISSFESGQIY 180
Query: 397 NHADAVGFIKLYGLPMRIRAMMEQGI 422
+ ADA GFI+LYGLP+R+RAM+EQGI
Sbjct: 181 DQADAAGFIRLYGLPVRVRAMLEQGI 258
>BM178846
Length = 421
Score = 126 bits (317), Expect = 2e-29
Identities = 64/83 (77%), Positives = 70/83 (84%), Gaps = 1/83 (1%)
Frame = +2
Query: 1 IKAVLYKDVEI-SETKKVAGLKGKLNKVVLAYSGGLDTSVVVPWLRENYGCDVVCFTADL 59
IKA D E+ SET K GL+GKLNKVVLAYSGGLDTSV+VPWLRENYGC+VVCFTAD+
Sbjct: 173 IKAAARIDTEVVSETTKGGGLRGKLNKVVLAYSGGLDTSVIVPWLRENYGCEVVCFTADV 352
Query: 60 GQGASELEGLEEKAKASGASQLV 82
GQG EL+GLE KAKASGA QLV
Sbjct: 353 GQGIKELDGLEAKAKASGACQLV 421
>AW459909
Length = 274
Score = 115 bits (288), Expect = 4e-26
Identities = 57/91 (62%), Positives = 69/91 (75%)
Frame = +2
Query: 113 TAIARPVIAKAMVDIANEVGADAVSHGCTGKGNDQVRFELSFFALNPKLNVVAPWREWDI 172
T++ P IAKAMVD+ E GADAV HGC+ KGNDQVRFE +FFA N N+ AP REW+I
Sbjct: 2 TSMCGPEIAKAMVDVGEEGGADAVCHGCSSKGNDQVRFEDTFFARNWMHNIAAPRREWNI 181
Query: 173 TGREDAIEYAKKHDVPVPVTKKSIYSRDRNL 203
T R + IEYAK H +PV +TK SIYSR+RN+
Sbjct: 182 TWR*NTIEYAKNHIMPVTMTKTSIYSRNRNI 274
>TC222133
Length = 319
Score = 29.6 bits (65), Expect = 2.7
Identities = 16/42 (38%), Positives = 21/42 (49%), Gaps = 6/42 (14%)
Frame = +2
Query: 332 ELVYAGRWFDPLRE------SMDSFMEKISQTTTGSVSLKLY 367
E+ Y WFDPL E S DS E+IS+ V ++ Y
Sbjct: 170 EICYPKGWFDPLEEPSLIE*SRDSISEEISKVNPSRVYIEKY 295
>AW306709 weakly similar to SP|P93329|NO20_ Early nodulin 20 precursor
(N-20). [Barrel medic] {Medicago truncatula}, partial
(8%)
Length = 414
Score = 29.3 bits (64), Expect = 3.5
Identities = 12/48 (25%), Positives = 27/48 (56%)
Frame = +3
Query: 175 REDAIEYAKKHDVPVPVTKKSIYSRDRNLWHITHEGDVLEDPANEPKK 222
+E+ + +K++ P PV++K D +WH+T+ G + ++ +K
Sbjct: 66 KENFLHCLRKNNRPYPVSRKE---DDSKIWHVTYMGPLFSRSSDPERK 200
>TC211813 similar to UP|Q9FH76 (Q9FH76) Cytochrome P450 monooxygenase
(AT5g45340/K9E15_12), partial (38%)
Length = 889
Score = 28.9 bits (63), Expect = 4.6
Identities = 18/62 (29%), Positives = 27/62 (43%), Gaps = 4/62 (6%)
Frame = +1
Query: 138 HGCTGKGNDQVRFELSFFALNPKLNVVAPWREWDITGREDAIEY----AKKHDVPVPVTK 193
H C G EL+ L L+ + W + G ED I+Y KH +PV +T
Sbjct: 427 HSCPGS-------ELAKLELLVLLHHLTLSYRWQVVGNEDGIQYGPFPVPKHGLPVKITP 585
Query: 194 KS 195
++
Sbjct: 586 RN 591
>CD404253
Length = 617
Score = 28.5 bits (62), Expect = 5.9
Identities = 17/41 (41%), Positives = 21/41 (50%)
Frame = -3
Query: 341 DPLRESMDSFMEKISQTTTGSVSLKLYKGSASVTGRKSPYS 381
D LR S E + T SV+ KG+ S TG+KSP S
Sbjct: 603 DLLRRSNSEGKESVVFLTPSSVNSAKKKGTKSETGKKSPAS 481
>TC230106 homologue to UP|O80412 (O80412) Mitochondrial phosphate
transporter, complete
Length = 1640
Score = 28.5 bits (62), Expect = 5.9
Identities = 16/39 (41%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Frame = +2
Query: 363 SLKLYKGSASVTGRKSPYSLYREDISSFES--GEIYNHA 399
+L+LYKG + GR+ PY++ + +SFE+ IY HA
Sbjct: 824 TLRLYKGLVPLWGRQIPYTMMK--FASFETIVELIYKHA 934
>TC217158 similar to UP|Q9SUW2 (Q9SUW2) Adenine phosphoribosyltransferase
-like protein , partial (95%)
Length = 1068
Score = 28.5 bits (62), Expect = 5.9
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 4/61 (6%)
Frame = +1
Query: 84 KDLKEEFVRDYVFPCLR--AGAIY--ERKYLLGTAIARPVIAKAMVDIANEVGADAVSHG 139
K + +E++ +Y CL GA+ ER ++ IA A +D+ VGA+ V G
Sbjct: 364 KVISQEYILEYGRDCLEMHVGAVEPGERALVVDDLIATGGTLCAAMDLLERVGAEVVECG 543
Query: 140 C 140
C
Sbjct: 544 C 546
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,251,915
Number of Sequences: 63676
Number of extensions: 177962
Number of successful extensions: 723
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 716
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 722
length of query: 422
length of database: 12,639,632
effective HSP length: 100
effective length of query: 322
effective length of database: 6,272,032
effective search space: 2019594304
effective search space used: 2019594304
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC126782.6


