

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126018.3 + phase: 0
(387 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
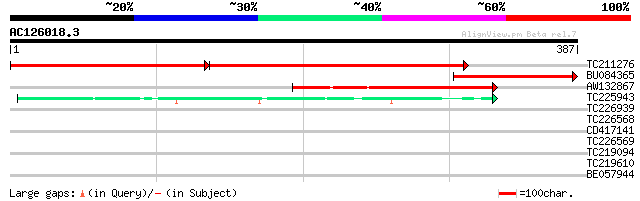
Score E
Sequences producing significant alignments: (bits) Value
TC211276 NADH dehydrogenase 341 3e-94
BU084365 160 1e-39
AW132867 homologue to PIR|T06260|T06 NADH2 dehydrogenase (ubiqui... 109 2e-24
TC225943 similar to UP|NU4C_LOTJA (Q9BBP3) NAD(P)H-quinone oxido... 56 3e-08
TC226939 similar to UP|Q9ZQW9 (Q9ZQW9) U2 snRNP auxiliary factor... 28 5.4
TC226568 28 7.0
CD417141 28 7.0
TC226569 28 7.0
TC219094 similar to PIR|T00565|T00565 geranylgeranyl-diphosphate... 28 7.0
TC219610 similar to UP|Q9STA2 (Q9STA2) Hydroperoxide lyase, part... 28 7.0
BE057944 28 9.2
>TC211276 NADH dehydrogenase
Length = 1632
Score = 341 bits (875), Expect = 3e-94
Identities = 171/177 (96%), Positives = 174/177 (97%)
Frame = +2
Query: 137 SPTPVVAFLSVTSKVAASASATRIFDILFYFSSNEWHLLLEILAILSMLLGNLIAITQTS 196
SPTPVVAFLSVTSKVAASASATRIFDI FYFSSNEWHLLLEILAILSM+LGNLIAITQTS
Sbjct: 1100 SPTPVVAFLSVTSKVAASASATRIFDIPFYFSSNEWHLLLEILAILSMILGNLIAITQTS 1279
Query: 197 MKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMNLGTFACIVLFGLRTGTD 256
MKRMLAYSSIGQIGYVIIGIIVGDSN GYASMITYMLFYISMNLGTFACIV FGLRTGTD
Sbjct: 1280 MKRMLAYSSIGQIGYVIIGIIVGDSNGGYASMITYMLFYISMNLGTFACIVSFGLRTGTD 1459
Query: 257 NIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWCGWQAGLYFLVSIG 313
NIRDYAGLYTKDP+LALSLALCLLSLGGLPPL+GFFGKLHLFWCGWQAGLYFLVSIG
Sbjct: 1460 NIRDYAGLYTKDPYLALSLALCLLSLGGLPPLSGFFGKLHLFWCGWQAGLYFLVSIG 1630
Score = 261 bits (666), Expect = 5e-70
Identities = 130/136 (95%), Positives = 131/136 (95%)
Frame = +1
Query: 1 MAITEFLLFILTATLGGMFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKY 60
MAITEFLLFILTATLGGMFLC ANDLITIFVALECFSLCSYLLSGYTKKDVRSNEAT KY
Sbjct: 1 MAITEFLLFILTATLGGMFLCGANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATTKY 180
Query: 61 LLMGGASSSILVHGFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIFITVGIGFK 120
LLMGGASSSILVHGFSWLYG SGGEIELQEIVNGLINTQMYNSPGI IAL+FITVGIGFK
Sbjct: 181 LLMGGASSSILVHGFSWLYGSSGGEIELQEIVNGLINTQMYNSPGILIALLFITVGIGFK 360
Query: 121 LSLAPSHQWTPDVYEG 136
LS APSHQWTPDVYEG
Sbjct: 361 LSPAPSHQWTPDVYEG 408
>BU084365
Length = 421
Score = 160 bits (404), Expect = 1e-39
Identities = 79/84 (94%), Positives = 83/84 (98%)
Frame = +2
Query: 304 AGLYFLVSIGLITSVVSIYYYLKIIKLLMTGRNQEITPHVRNYRRSPVRSNNSIELSMII 363
AGLYFLVSIGL+TSVVSIYYYLKIIKLLMTGRNQEITPHVRNYRR P+RSNNSIELSMI+
Sbjct: 2 AGLYFLVSIGLLTSVVSIYYYLKIIKLLMTGRNQEITPHVRNYRRPPLRSNNSIELSMIV 181
Query: 364 CVIASTVPGISMNPIIEIAQDTLF 387
CVIAST+PGISMNPIIEIAQDTLF
Sbjct: 182 CVIASTIPGISMNPIIEIAQDTLF 253
>AW132867 homologue to PIR|T06260|T06 NADH2 dehydrogenase (ubiquinone) (EC
1.6.5.3) chain 2 - wheat mitochondrion, partial (28%)
Length = 420
Score = 109 bits (272), Expect = 2e-24
Identities = 56/140 (40%), Positives = 87/140 (62%)
Frame = -1
Query: 194 QTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMNLGTFACIVLFGLRT 253
QT +KR+LAYSSIG +GY+ IG G + +G S++ +L Y M + FA IVL +T
Sbjct: 417 QTKVKRLLAYSSIGHVGYICIGFSCG-TIEGIQSLLIGILIYALMTIDAFA-IVLALRQT 244
Query: 254 GTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWCGWQAGLYFLVSIG 313
I D L +P LA++ ++ + S G+PPLAGF K +LF+ G YFL S+G
Sbjct: 243 RVKYIADLGALAKTNPILAITFSITMFSYVGIPPLAGFCSKFYLFFAALGCGAYFLASVG 64
Query: 314 LITSVVSIYYYLKIIKLLMT 333
++TSV+ +YY++++K + +
Sbjct: 63 VVTSVIGCFYYIRLVKRMFS 4
>TC225943 similar to UP|NU4C_LOTJA (Q9BBP3) NAD(P)H-quinone oxidoreductase
chain 4, chloroplast (NAD(P)H dehydrogenase, chain 4)
(NADH-plastoquinone oxidoreductase chain 4) , partial
(86%)
Length = 1432
Score = 55.8 bits (133), Expect = 3e-08
Identities = 76/339 (22%), Positives = 137/339 (39%), Gaps = 11/339 (3%)
Frame = +1
Query: 6 FLLFILTATLGGMFLCSANDLITIFVALECFSLCSYLLSGYTKKDVRSNEATMKYLLMGG 65
F +L G + + S+ D++ F+ E + YLL R AT K++L
Sbjct: 364 FYFLMLAMYSGQIGIFSSQDILLFFIMWEFELIPVYLLLSMWGGKKRLYSAT-KFILYTA 540
Query: 66 ASSSILVHGFSWLYGLSGGEIELQEIVNGLINTQMYNSPGISIALIF---ITVGIGFKLS 122
SS L+ G + S E L L N S +++ +IF + K
Sbjct: 541 GSSVFLLLGILGMSFYSSNEPTLN--FESLTN----QSYPVALEIIFYIGFLIAFAVKSP 702
Query: 123 LAPSHQWTPDVY-EGSPTPVVAFLSVTSKVAASASATRIFDILFYFSS--NEWHLLLEIL 179
+ P H W PD + E + + + K+ A + L S + W L +L
Sbjct: 703 IIPLHTWLPDTHGEAHYSTCMLLAGILLKMGAYGLVRINMEFLSRAHSIFSPW---LILL 873
Query: 180 AILSMLLGNLIAITQTSMKRMLAYSSIGQIGYVIIGIIVGDSNDGYASMITYMLFYISMN 239
+ ++ ++ Q ++K+ +AYSS+ +G++++G I S+ G I ++ +
Sbjct: 874 GSIQIIYAASTSLGQRNLKKRIAYSSVSHMGFLLLG-IGSISDTGLNGAILQIISH---- 1038
Query: 240 LGTFACIVLFGLRTGTDNIR-----DYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGK 294
G + F T D +R + G+ P + + ++ LP ++GF +
Sbjct: 1039-GFIGAALFFLAGTSYDRLRLLYLDEMGGMAIPMPKIFTVFTILSMASLALPGMSGFVAE 1215
Query: 295 LHLFWCGWQAGLYFLVSIGLITSVVSIYYYLKIIKLLMT 333
L +V +G+ITS YL I K+L+T
Sbjct: 1216L-------------IVLLGIITS----QKYLLITKILIT 1281
>TC226939 similar to UP|Q9ZQW9 (Q9ZQW9) U2 snRNP auxiliary factor, small
subunit, partial (74%)
Length = 1397
Score = 28.5 bits (62), Expect = 5.4
Identities = 19/43 (44%), Positives = 27/43 (62%), Gaps = 2/43 (4%)
Frame = -1
Query: 149 SKVAASASATRIFDILFYFSSNEWHLLLEILAILSML--LGNL 189
S +AA+A+ R +I F FSSN LLL +L +L + +GNL
Sbjct: 161 SAIAAAATTHRSNEIRFDFSSNSSLLLLLLLLLLPRVCEVGNL 33
>TC226568
Length = 946
Score = 28.1 bits (61), Expect = 7.0
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = +2
Query: 298 FWCGWQAGLYFLVSIGLIT 316
FWC Q L+FL+S+GL +
Sbjct: 302 FWCRLQRTLFFLISLGLFS 358
>CD417141
Length = 691
Score = 28.1 bits (61), Expect = 7.0
Identities = 15/24 (62%), Positives = 16/24 (66%)
Frame = -1
Query: 248 LFGLRTGTDNIRDYAGLYTKDPFL 271
LFG TG N+ DY GL TK PFL
Sbjct: 322 LFGWGTGQ*NVLDYGGLTTK-PFL 254
>TC226569
Length = 1432
Score = 28.1 bits (61), Expect = 7.0
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = +1
Query: 298 FWCGWQAGLYFLVSIGLIT 316
FWC Q L+FL+S+GL +
Sbjct: 757 FWCRLQRTLFFLISLGLFS 813
>TC219094 similar to PIR|T00565|T00565 geranylgeranyl-diphosphate
geranylgeranyltransferase I beta chain homolog At2g39550
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(36%)
Length = 456
Score = 28.1 bits (61), Expect = 7.0
Identities = 20/69 (28%), Positives = 35/69 (49%)
Frame = +2
Query: 253 TGTDNIRDYAGLYTKDPFLALSLALCLLSLGGLPPLAGFFGKLHLFWCGWQAGLYFLVSI 312
TG+ R+ G++ D ++LA+ ++ G L P GF+G +L FLV+I
Sbjct: 155 TGSCRGRELMGVFKVDQ---INLAIHVMHFG-LEPF*GFWGASNLLTIRLYVDFCFLVNI 322
Query: 313 GLITSVVSI 321
++ S S+
Sbjct: 323 SMVVSANSL 349
>TC219610 similar to UP|Q9STA2 (Q9STA2) Hydroperoxide lyase, partial (51%)
Length = 815
Score = 28.1 bits (61), Expect = 7.0
Identities = 27/73 (36%), Positives = 36/73 (48%)
Frame = +2
Query: 226 ASMITYMLFYISMNLGTFACIVLFGLRTGTDNIRDYAGLYTKDPFLALSLALCLLSLGGL 285
+S ++Y LF + L TF C VL G D +G + +LAL L L +S+G L
Sbjct: 560 SSSVSY-LFPLQQFLFTFLCKVLAGADPARDPKIAESGYSMLNSWLALQL-LPTVSVGIL 733
Query: 286 PPLAGFFGKLHLF 298
PL F LH F
Sbjct: 734 QPLEEIF--LHSF 766
>BE057944
Length = 279
Score = 27.7 bits (60), Expect = 9.2
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Frame = +2
Query: 278 CLLSLG-GLPPLAGFFGKLHLFWCGW 302
C L++G GLPP GF+ LF GW
Sbjct: 131 CSLNVGFGLPPCLGFYVCFTLFGVGW 208
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,346,159
Number of Sequences: 63676
Number of extensions: 251407
Number of successful extensions: 1855
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 1841
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1854
length of query: 387
length of database: 12,639,632
effective HSP length: 99
effective length of query: 288
effective length of database: 6,335,708
effective search space: 1824683904
effective search space used: 1824683904
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC126018.3


