

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126012.1 + phase: 0
(220 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
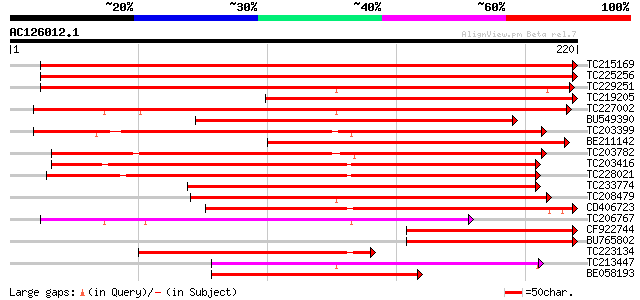
Score E
Sequences producing significant alignments: (bits) Value
TC215169 UP|6DCS_SOYBN (P26690) NAD(P)H dependent 6'-deoxychalco... 363 e-101
TC225256 weakly similar to UP|6DCS_SOYBN (P26690) NAD(P)H depend... 310 4e-85
TC229251 similar to UP|Q40333 (Q40333) Chalcone reductase , part... 270 4e-73
TC219205 homologue to UP|Q8S4C1 (Q8S4C1) Chalcone reductase, par... 216 7e-57
TC227002 similar to UP|Q41399 (Q41399) Chalcone reductase, parti... 208 1e-54
BU549390 similar to GP|20147510|gb chalcone reductase {Pueraria ... 197 4e-51
TC203399 similar to UP|Q41399 (Q41399) Chalcone reductase, complete 193 6e-50
BE211142 similar to SP|P26690|6DCS NAD(P)H dependent 6'-deoxycha... 171 3e-43
TC203782 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (97%) 166 1e-41
TC203416 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (87%) 164 3e-41
TC228021 similar to PIR|T02543|T02543 aldehyde dehydrogenase hom... 155 1e-38
TC233774 similar to UP|Q9V9A4 (Q9V9A4) CG9436-PA (LD24696p), par... 125 2e-29
TC208479 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (50%) 120 6e-28
CD406723 similar to PIR|T09670|T09 abscisic acid activated prote... 111 2e-25
TC206767 similar to UP|Q9FVN7 (Q9FVN7) NADPH-dependent mannose 6... 108 2e-24
CF922744 102 1e-22
BU765802 102 1e-22
TC223134 weakly similar to UP|GCY_YEAST (P14065) GCY protein , ... 85 2e-17
TC213447 weakly similar to GB|AAC53199.1|2114406|CGU81045 aldo-k... 81 4e-16
BE058193 weakly similar to GP|29028836|gb At2g37790 {Arabidopsis... 75 3e-14
>TC215169 UP|6DCS_SOYBN (P26690) NAD(P)H dependent 6'-deoxychalcone synthase
, complete
Length = 1517
Score = 363 bits (932), Expect = e-101
Identities = 175/208 (84%), Positives = 193/208 (92%)
Frame = +3
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S +TLQL+YLDLYLIHWPLSSQPGKF+FPI+V DLLPFDVKGVWESMEE KLGLTKAIG
Sbjct: 675 SLKTLQLEYLDLYLIHWPLSSQPGKFSFPIEVEDLLPFDVKGVWESMEECQKLGLTKAIG 854
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRG 132
VSNFSVKKL+NLLSVATI P V+QVEMNLAWQQKKLREFC NGI++TAFSPLRKGASRG
Sbjct: 855 VSNFSVKKLQNLLSVATIRPVVDQVEMNLAWQQKKLREFCKENGIIVTAFSPLRKGASRG 1034
Query: 133 PNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKE 192
PNEVMEND+LKEIA+AHGKS+AQ+SLRWLYEQGVTFVPKSYDKERMNQNL IFDW+LT++
Sbjct: 1035PNEVMENDVLKEIAEAHGKSIAQVSLRWLYEQGVTFVPKSYDKERMNQNLHIFDWALTEQ 1214
Query: 193 DHEKIDQIKQNRLIPGPTKPGINDLYDD 220
DH KI QI Q+RLI GPTKP + DL+DD
Sbjct: 1215DHHKISQISQSRLISGPTKPQLADLWDD 1298
>TC225256 weakly similar to UP|6DCS_SOYBN (P26690) NAD(P)H dependent
6'-deoxychalcone synthase , partial (95%)
Length = 1196
Score = 310 bits (793), Expect = 4e-85
Identities = 145/208 (69%), Positives = 181/208 (86%)
Frame = +3
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S RTLQL+Y+DL+LIHWP++++PGK +PI+V++++ FD+KGVW SMEE +LGLTKAIG
Sbjct: 348 SLRTLQLEYIDLFLIHWPIATKPGKVVYPIEVSEIVEFDMKGVWGSMEECQRLGLTKAIG 527
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRG 132
VSNFS+KKLE LLS ATI PAVNQVE+NL WQQ+KLR+FC GI +TAFSPLRKGASRG
Sbjct: 528 VSNFSIKKLEKLLSFATIPPAVNQVEVNLGWQQQKLRDFCKEKGITVTAFSPLRKGASRG 707
Query: 133 PNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKE 192
N V++ND++KE+ADAHGK+ AQI LRWLYEQG+TFV KSYDKERM QNL IFDWSLT++
Sbjct: 708 ANFVLDNDVIKELADAHGKTAAQICLRWLYEQGLTFVVKSYDKERMKQNLGIFDWSLTED 887
Query: 193 DHEKIDQIKQNRLIPGPTKPGINDLYDD 220
D++KI +I Q RLI GPTKP ++DL+D+
Sbjct: 888 DYKKISEIHQERLIKGPTKPLLDDLWDE 971
>TC229251 similar to UP|Q40333 (Q40333) Chalcone reductase , partial (53%)
Length = 934
Score = 270 bits (690), Expect = 4e-73
Identities = 129/209 (61%), Positives = 167/209 (79%), Gaps = 2/209 (0%)
Frame = +1
Query: 13 SCRTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIG 72
S R+L+LDYLDLYLIHWP++++PG + P L+PFD+K VW +MEE KLGLTK+IG
Sbjct: 22 SLRSLKLDYLDLYLIHWPITAKPGMWEMPYSEESLVPFDLKSVWAAMEECHKLGLTKSIG 201
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGASR 131
VSNFS KKLENLLS ATI P+VNQVEMN+AWQQK LR +C A GI++TA+SPL KG+
Sbjct: 202 VSNFSCKKLENLLSFATIPPSVNQVEMNIAWQQKNLRAYCKAKGIIVTAYSPLGAKGSKW 381
Query: 132 GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTK 191
N++++N++ K+IA AHGK+ AQ+ LRWL+EQGVTF+PKSY+KER+ +NL IFDWSLTK
Sbjct: 382 DINQILDNELTKQIAQAHGKTAAQVCLRWLFEQGVTFIPKSYNKERLKENLEIFDWSLTK 561
Query: 192 EDHEKIDQIKQNRLIP-GPTKPGINDLYD 219
+DHEKI+Q+KQ R+ G + DL+D
Sbjct: 562 DDHEKINQVKQERMFKYGTAAFPLPDLFD 648
>TC219205 homologue to UP|Q8S4C1 (Q8S4C1) Chalcone reductase, partial (39%)
Length = 515
Score = 216 bits (550), Expect = 7e-57
Identities = 100/121 (82%), Positives = 113/121 (92%)
Frame = +1
Query: 100 NLAWQQKKLREFCNANGIVLTAFSPLRKGASRGPNEVMENDMLKEIADAHGKSVAQISLR 159
+LAWQQKKLREFC NGI+LTAFSPLRKGAS+GPNEVMEND+LKEIA+AHGKS+AQ+SLR
Sbjct: 7 SLAWQQKKLREFCKENGIILTAFSPLRKGASKGPNEVMENDVLKEIAEAHGKSIAQVSLR 186
Query: 160 WLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPGPTKPGINDLYD 219
WLYEQGVTFVPKSYDKERMNQNL IFDW+LT+EDH KID+I Q+RLI GPTKP + DL+D
Sbjct: 187 WLYEQGVTFVPKSYDKERMNQNLQIFDWALTEEDHHKIDEIYQSRLISGPTKPQVTDLWD 366
Query: 220 D 220
D
Sbjct: 367 D 369
>TC227002 similar to UP|Q41399 (Q41399) Chalcone reductase, partial (93%)
Length = 1165
Score = 208 bits (530), Expect = 1e-54
Identities = 114/212 (53%), Positives = 148/212 (69%), Gaps = 3/212 (1%)
Frame = +3
Query: 10 LLISCRTLQLDYLDLYLIHWPLSSQP-GKFTFPIDVADLLP-FDVKGVWESMEEGLKLGL 67
L S + L L+Y+DLYLIHWP+ +P K I ++LP FD+KG+WE+MEE +LGL
Sbjct: 303 LKTSLQKLGLEYVDLYLIHWPVRLKPEAKGYHNILKENVLPSFDMKGIWEAMEECYRLGL 482
Query: 68 TKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-R 126
K+IGVSNF +KKL LL ATI PAVNQVEM+ AWQQ KL+EFC GI ++A+SPL
Sbjct: 483 AKSIGVSNFGIKKLSQLLENATIPPAVNQVEMSPAWQQGKLKEFCKQKGIHVSAWSPLGA 662
Query: 127 KGASRGPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFD 186
+++G N VME+ +LKEIA KS+AQI+LRW+YEQG + KS++KERM QNL IFD
Sbjct: 663 YKSAQGTNAVMESPILKEIACERQKSMAQIALRWIYEQGAIAIVKSFNKERMKQNLDIFD 842
Query: 187 WSLTKEDHEKIDQIKQNRLIPGPTKPGINDLY 218
W L++E+ +K QI Q R+ G T N Y
Sbjct: 843 WELSQEESQKFSQIPQRRMYRGITFVSENGPY 938
>BU549390 similar to GP|20147510|gb chalcone reductase {Pueraria montana var.
lobata}, partial (40%)
Length = 517
Score = 197 bits (500), Expect = 4e-51
Identities = 94/125 (75%), Positives = 110/125 (87%)
Frame = -3
Query: 73 VSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRG 132
VSNFS+KKLE LLS ATI PAVNQVE+NL WQQ+KLR+FC GI +TAFSPLRKGASRG
Sbjct: 515 VSNFSIKKLEKLLSFATIPPAVNQVEVNLGWQQQKLRDFCKEKGITVTAFSPLRKGASRG 336
Query: 133 PNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKE 192
N V++ND++KE+ADAHGK+ AQI LRWLYEQG+TFV KSYDKERM QNL IFDWSLT++
Sbjct: 335 ANFVLDNDVIKELADAHGKTAAQICLRWLYEQGLTFVVKSYDKERMKQNLGIFDWSLTED 156
Query: 193 DHEKI 197
D++KI
Sbjct: 155 DYKKI 141
>TC203399 similar to UP|Q41399 (Q41399) Chalcone reductase, complete
Length = 1148
Score = 193 bits (490), Expect = 6e-50
Identities = 103/207 (49%), Positives = 141/207 (67%), Gaps = 8/207 (3%)
Frame = +1
Query: 10 LLISCRTLQLDYLDLYLIHWPLS-----SQPGKFTFPIDVADLLPFDVKGVWESMEEGLK 64
L + + L +Y+DLYLIHWP+ P FT D+LPFD++G W++MEE K
Sbjct: 331 LKTTLKKLGTEYVDLYLIHWPVRLRHDLENPTVFT----KEDVLPFDIEGTWKAMEECYK 498
Query: 65 LGLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSP 124
LG+ K+IG+ N+ +KKL LL +ATI PAVNQVEMN +WQQ KLREFC GI ++A+S
Sbjct: 499 LGIAKSIGICNYGIKKLTKLLEIATIPPAVNQVEMNPSWQQGKLREFCKQKGIHVSAWSA 678
Query: 125 LRKGASR---GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQN 181
L GA + G VMEN +L++IA A GK++AQ++LRW+Y+QG + + KS + ERM QN
Sbjct: 679 L--GAYKIFWGSGAVMENPILQDIAKAKGKTIAQVALRWVYQQGSSAMAKSTNSERMKQN 852
Query: 182 LCIFDWSLTKEDHEKIDQIKQNRLIPG 208
L IFD+ L++ED E+I Q+ Q R G
Sbjct: 853 LDIFDFVLSEEDLERISQVPQRRQYTG 933
>BE211142 similar to SP|P26690|6DCS NAD(P)H dependent 6'-deoxychalcone
synthase (EC 1.1.-.-). [Soybean] {Glycine max}, partial
(36%)
Length = 352
Score = 171 bits (432), Expect = 3e-43
Identities = 83/117 (70%), Positives = 99/117 (83%)
Frame = +2
Query: 101 LAWQQKKLREFCNANGIVLTAFSPLRKGASRGPNEVMENDMLKEIADAHGKSVAQISLRW 160
LAWQQKKLRE C NGI++TAFSPLRKGASRGPNEVMEND+LKEIA+AHGKS+AQ+SLR
Sbjct: 2 LAWQQKKLRELCKENGIIVTAFSPLRKGASRGPNEVMENDVLKEIAEAHGKSIAQMSLR* 181
Query: 161 LYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPGPTKPGINDL 217
LY+Q VTF+PK YD+E MNQNL IF+ +LTK+DH KI QI + +I PTKP + +L
Sbjct: 182 LYKQSVTFMPKIYDEEIMNQNLHIFN*ALTKQDHHKISQISHSHVIS*PTKPQLTNL 352
>TC203782 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (97%)
Length = 1490
Score = 166 bits (419), Expect = 1e-41
Identities = 86/194 (44%), Positives = 128/194 (65%), Gaps = 2/194 (1%)
Frame = +3
Query: 17 LQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNF 76
LQLDYLDLYLIHWP+ + G F + D D+ W++ME G +AIGVSNF
Sbjct: 474 LQLDYLDLYLIHWPVRMKSGSVGFKKEYLDQP--DIPSTWKAMEALYDSGKARAIGVSNF 647
Query: 77 SVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRG--PN 134
S KKL++L+++A + PAVNQVE++ WQQ KL FC + G+ L+ +SPL S G +
Sbjct: 648 SSKKLQDLMNIARVPPAVNQVELHPGWQQPKLHAFCESKGVHLSGYSPL---GSPGVLKS 818
Query: 135 EVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDH 194
++++N ++ EIA+ GK+ AQ++LRW + G + +PKS ++ R+ N +FDWS+ +E
Sbjct: 819 DILKNPVVIEIAEKLGKTPAQVALRWGLQTGHSVLPKSTNESRIKGNFDVFDWSIPEEVM 998
Query: 195 EKIDQIKQNRLIPG 208
+K +IKQ+RLI G
Sbjct: 999 DKFSEIKQDRLIKG 1040
>TC203416 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (87%)
Length = 1225
Score = 164 bits (415), Expect = 3e-41
Identities = 86/190 (45%), Positives = 126/190 (66%)
Frame = +1
Query: 17 LQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVSNF 76
LQLDYLDLYLIHWP+ + G TF ++ L D+ W +ME +AIGVSNF
Sbjct: 322 LQLDYLDLYLIHWPVRMKSG--TFGLNKEYLEQPDIPNTWRAMEALYDSDKARAIGVSNF 495
Query: 77 SVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRGPNEV 136
S KKL++LL +A ++PAVNQVE++ WQQ KLR FC + I L+ +SPL A+ +++
Sbjct: 496 SSKKLQDLLDIARVVPAVNQVELHPGWQQPKLRAFCESKEIHLSGYSPLGSPAAL-KSDI 672
Query: 137 MENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEK 196
++N ++ EIA+ GK+ AQ++LRW + G + +PKS ++ R+ N IFDWS+ ++ K
Sbjct: 673 LKNPVVTEIAERLGKTPAQVALRWGLQAGHSVLPKSTNESRIKGNFDIFDWSIPQDLMTK 852
Query: 197 IDQIKQNRLI 206
I +IKQ RL+
Sbjct: 853 ISEIKQERLV 882
>TC228021 similar to PIR|T02543|T02543 aldehyde dehydrogenase homolog
At2g37770 - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (98%)
Length = 1119
Score = 155 bits (392), Expect = 1e-38
Identities = 80/192 (41%), Positives = 120/192 (61%)
Frame = +3
Query: 15 RTLQLDYLDLYLIHWPLSSQPGKFTFPIDVADLLPFDVKGVWESMEEGLKLGLTKAIGVS 74
R LQLDY+DLYLIHWP+ + G F + +++P D+ W++ME K G +AIGVS
Sbjct: 297 RDLQLDYIDLYLIHWPIRMKKGSVGFKAE--NIVPSDIPNTWKAMEALNKSGKARAIGVS 470
Query: 75 NFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRGPN 134
NFS KKL LL A + PAVNQ E + AW+Q KL+ FC + G+ + +SPL A
Sbjct: 471 NFSTKKLGELLEYARVTPAVNQSECHPAWRQDKLKAFCKSKGVHFSGYSPLGSPAWL-EG 647
Query: 135 EVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDH 194
+ + + ++ IA GK+ AQ++LRW + G + +PKS + R+ +N IFDWS+ ++
Sbjct: 648 DFLNHPVINMIAKKLGKTPAQVALRWGLQMGHSVLPKSSNPARIKENFDIFDWSIPEDML 827
Query: 195 EKIDQIKQNRLI 206
+K +I+Q RL+
Sbjct: 828 DKFFEIQQERLL 863
>TC233774 similar to UP|Q9V9A4 (Q9V9A4) CG9436-PA (LD24696p), partial (7%)
Length = 447
Score = 125 bits (313), Expect = 2e-29
Identities = 64/137 (46%), Positives = 88/137 (63%)
Frame = +2
Query: 70 AIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGA 129
AIGVSNFS+KKL++LL A I PAVNQVE + WQQ L C + G+ LTA+ PL
Sbjct: 14 AIGVSNFSIKKLQDLLGYAKIPPAVNQVECHPVWQQPALHNLCKSTGVHLTAYCPLGSPG 193
Query: 130 SRGPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSL 189
S E+++ +L EIA+ KS AQ++LRW + G + +PKS ++ R+ +NL +FDW L
Sbjct: 194 SWVKGEILKEPLLIEIAEKLHKSPAQVALRWGLQSGHSVLPKSVNESRIKENLSLFDWCL 373
Query: 190 TKEDHEKIDQIKQNRLI 206
E K+ QI Q RL+
Sbjct: 374 PPELFSKLSQIHQQRLL 424
>TC208479 similar to UP|Q84TF0 (Q84TF0) At2g37790, partial (50%)
Length = 810
Score = 120 bits (300), Expect = 6e-28
Identities = 60/141 (42%), Positives = 96/141 (67%), Gaps = 1/141 (0%)
Frame = +1
Query: 71 IGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL-RKGA 129
IGVSNFS KKL +LL +A + PAVNQVE + +WQQ KL+ FCN+ G+ L+ +SPL G
Sbjct: 7 IGVSNFSTKKLSDLLLIARVPPAVNQVECHPSWQQDKLQAFCNSKGVHLSGYSPLGSPGT 186
Query: 130 SRGPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSL 189
+ ++V+++ ++ IA+ GK+ AQ++LRW + G + +PKS ++ R+ +N +F WS+
Sbjct: 187 TWLKSDVLKHQVINMIAEKLGKTPAQVALRWGLQMGHSVLPKSTNETRIKENFDVFGWSI 366
Query: 190 TKEDHEKIDQIKQNRLIPGPT 210
++ K +I+Q RL+ G T
Sbjct: 367 PEDLLAKFSEIQQARLLRGTT 429
>CD406723 similar to PIR|T09670|T09 abscisic acid activated protein -
alfalfa, partial (38%)
Length = 691
Score = 111 bits (278), Expect = 2e-25
Identities = 61/152 (40%), Positives = 94/152 (61%), Gaps = 8/152 (5%)
Frame = -3
Query: 77 SVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRGPNEV 136
SVKKL++LL VA++ PAVNQVE++ + QQ +L FC + G+ L+ +SPL KG S + +
Sbjct: 689 SVKKLQDLLDVASVPPAVNQVELHPSLQQPELHAFCKSKGVHLSGYSPLGKGYSE--SNI 516
Query: 137 MENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEK 196
++N L A+ GK+ AQI+LRW + G + +PKS + R+ +N +FDWS+ +
Sbjct: 515 LKNPFLHTTAEKLGKTAAQIALRWGLQMGHSVLPKSTNDARLKENFDLFDWSIPADLLAN 336
Query: 197 IDQIKQNRLIPG-----PTKPG---INDLYDD 220
IKQ R++ G T PG I +L+D+
Sbjct: 335 FSDIKQERIVTGDGFFSKTSPGYKTIEELWDE 240
>TC206767 similar to UP|Q9FVN7 (Q9FVN7) NADPH-dependent mannose 6-phosphate
reductase, complete
Length = 1323
Score = 108 bits (269), Expect = 2e-24
Identities = 61/179 (34%), Positives = 105/179 (58%), Gaps = 11/179 (6%)
Frame = +3
Query: 13 SCRTLQLDYLDLYLIHWPLSSQP---GKFTFPIDVADLLPFD----VKGVWESMEEGLKL 65
S + LQL YLDLYL+H+P++ + G + P+ +L D ++ W +ME+ +
Sbjct: 315 SLKKLQLTYLDLYLVHFPVAVRHTGVGNTSSPLGDDGVLDIDTTISLETTWHAMEDLVSS 494
Query: 66 GLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL 125
GL ++IG+SN+ + + L+ + I PAVNQ+E + +Q+ L +FC +GI +TA +PL
Sbjct: 495 GLVRSIGISNYDIFLTRDCLAYSKIKPAVNQIETHPYFQRDSLVKFCQKHGICVTAHTPL 674
Query: 126 RKGASR----GPNEVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQ 180
A+ G +++ +LK +A+ + K+ AQISLRW ++ +PKS ER+ +
Sbjct: 675 GGAAANAEWFGTVSCLDDQVLKGLAEKYKKTAAQISLRWGIQRNTVVIPKSSKLERLKR 851
>CF922744
Length = 497
Score = 102 bits (255), Expect = 1e-22
Identities = 47/66 (71%), Positives = 57/66 (86%)
Frame = +3
Query: 155 QISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPGPTKPGI 214
QI LRWLYEQG+TFV KSYDKERM QNL IFDWSLT++D++KI +I Q RLI GPTKP +
Sbjct: 102 QICLRWLYEQGLTFVVKSYDKERMKQNLGIFDWSLTEDDYKKISEIHQERLIKGPTKPLL 281
Query: 215 NDLYDD 220
+DL+D+
Sbjct: 282 DDLWDE 299
>BU765802
Length = 396
Score = 102 bits (255), Expect = 1e-22
Identities = 47/66 (71%), Positives = 57/66 (86%)
Frame = +2
Query: 155 QISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDHEKIDQIKQNRLIPGPTKPGI 214
QI LRWLYEQG+TFV KSYDKERM QNL IFDWSLT++D++KI +I Q RLI GPTKP +
Sbjct: 2 QICLRWLYEQGLTFVVKSYDKERMKQNLGIFDWSLTEDDYKKISEIHQERLIKGPTKPLL 181
Query: 215 NDLYDD 220
+DL+D+
Sbjct: 182 DDLWDE 199
>TC223134 weakly similar to UP|GCY_YEAST (P14065) GCY protein , partial
(10%)
Length = 627
Score = 85.1 bits (209), Expect = 2e-17
Identities = 43/92 (46%), Positives = 62/92 (66%)
Frame = +2
Query: 51 DVKGVWESMEEGLKLGLTKAIGVSNFSVKKLENLLSVATILPAVNQVEMNLAWQQKKLRE 110
D+ W +ME G +AIGVSNFSVKKL++LL VA++ PAVNQVE++ + QQ +L
Sbjct: 41 DIPSTWRAMEALYNSGKAQAIGVSNFSVKKLQDLLDVASVPPAVNQVELHPSLQQPELHA 220
Query: 111 FCNANGIVLTAFSPLRKGASRGPNEVMENDML 142
FC + G+ L+ +SPL KG S + +++N L
Sbjct: 221 FCKSKGVHLSGYSPLGKGYSE--SNILKNPFL 310
>TC213447 weakly similar to GB|AAC53199.1|2114406|CGU81045 aldo-keto
reductase {Cricetulus griseus;} , partial (39%)
Length = 567
Score = 80.9 bits (198), Expect = 4e-16
Identities = 46/134 (34%), Positives = 75/134 (55%), Gaps = 5/134 (3%)
Frame = +1
Query: 79 KKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPL----RKGASRGPN 134
++L+ LL VA I P NQ+E++ Q KL +C +++TA+SP R A
Sbjct: 1 EQLDRLLKVAKIKPVTNQIEVHPYLNQSKLISYCKERDVLVTAYSPFASPDRPWAKPTDP 180
Query: 135 EVMENDMLKEIADAHGKSVAQISLRWLYEQGVTFVPKSYDKERMNQNLCIFDWSLTKEDH 194
++++ ++ IAD H KS A + LR+L ++G +PKS R+ N +FD+ LT E+
Sbjct: 181 SLLDDPIIISIADKHKKSNAHVILRYLIQRGTVPIPKSVTPSRIQTNFQVFDFELTAEEI 360
Query: 195 EKIDQIKQN-RLIP 207
+ ID + N R+ P
Sbjct: 361 KTIDGLDCNGRICP 402
>BE058193 weakly similar to GP|29028836|gb At2g37790 {Arabidopsis thaliana},
partial (35%)
Length = 367
Score = 74.7 bits (182), Expect = 3e-14
Identities = 38/82 (46%), Positives = 52/82 (63%)
Frame = +2
Query: 79 KKLENLLSVATILPAVNQVEMNLAWQQKKLREFCNANGIVLTAFSPLRKGASRGPNEVME 138
+KL++LL A I PAVNQVE + WQQ L C + G+ LTA+ PL S +V++
Sbjct: 5 EKLQDLLGYAKIPPAVNQVECHPVWQQPALHNLCKSTGVHLTAYCPLGSPGSWVKGQVLK 184
Query: 139 NDMLKEIADAHGKSVAQISLRW 160
+LKEIA+ KS AQ++LRW
Sbjct: 185 EPLLKEIAEKLHKSPAQVALRW 250
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,916,756
Number of Sequences: 63676
Number of extensions: 127364
Number of successful extensions: 668
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 648
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 650
length of query: 220
length of database: 12,639,632
effective HSP length: 93
effective length of query: 127
effective length of database: 6,717,764
effective search space: 853156028
effective search space used: 853156028
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC126012.1


