

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126009.2 + phase: 0
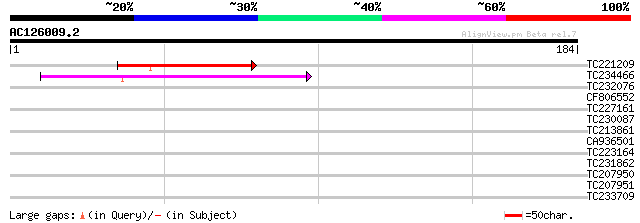
(184 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221209 44 4e-05
TC234466 41 3e-04
TC232076 weakly similar to UP|Q9ZVC6 (Q9ZVC6) Expressed protein,... 39 0.001
CF806552 33 0.076
TC227161 similar to UP|Q9LUM0 (Q9LUM0) Emb|CAB36798.1, partial (... 31 0.29
TC230087 similar to GB|BAB08313.1|9757977|AB018107 heat shock hs... 30 0.64
TC213861 28 2.4
CA936501 28 2.4
TC223164 27 5.5
TC231862 similar to GB|AAL60196.1|18139887|AF441079 O-linked N-a... 27 5.5
TC207950 similar to GB|AAT06459.1|46931310|BT012640 At3g57910 {A... 27 7.1
TC207951 similar to GB|AAT06459.1|46931310|BT012640 At3g57910 {A... 27 7.1
TC233709 26 9.3
>TC221209
Length = 939
Score = 43.9 bits (102), Expect = 4e-05
Identities = 19/47 (40%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Frame = +2
Query: 36 NGERPFDGT--KWKRFKKEFELPKYCNEDAIRGNFMQNILSVVLPKK 80
+GER D + KW RF+KE ++ K CN ++IR F +L + +PK+
Sbjct: 557 HGERLVDASNDKWSRFRKEIKISKGCNMNSIRAKFSHGVLFIAMPKE 697
>TC234466
Length = 780
Score = 41.2 bits (95), Expect = 3e-04
Identities = 22/107 (20%), Positives = 46/107 (42%), Gaps = 19/107 (17%)
Frame = +1
Query: 11 YEDFDPVFKWRREQDRDTIELHLPG-------------------NGERPFDGTKWKRFKK 51
YE F+P + + +++ + ++LPG GERP G + F++
Sbjct: 118 YETFEPKSEMKEKEEAYFLHIYLPGFVKEKIKINFVGSSRVVRVAGERPLGGNRISNFEQ 297
Query: 52 EFELPKYCNEDAIRGNFMQNILSVVLPKKVDLIPQEEQEEEEKIPEL 98
+ +P+ C ++G + L + +PKK + + + E P +
Sbjct: 298 AYPVPENCEVGKLQGKYELGTLIITMPKKPIISQVSPKAKVETTPRI 438
>TC232076 weakly similar to UP|Q9ZVC6 (Q9ZVC6) Expressed protein, partial
(18%)
Length = 997
Score = 38.9 bits (89), Expect = 0.001
Identities = 28/112 (25%), Positives = 48/112 (42%), Gaps = 23/112 (20%)
Frame = +3
Query: 6 AHNRSYEDFDPVFKWRREQ----------DRDTIELHLPGN------GERPFDGTKWKRF 49
A +R YEDF P+++W R + RD +++ + GER +W+RF
Sbjct: 69 ASDRVYEDFVPLYEWDRNERLVNVMLPGFRRDQLKVQVTSKLTLRLMGERLITDNRWRRF 248
Query: 50 KKEFELPKYCNEDAIRGNFMQNILSV-------VLPKKVDLIPQEEQEEEEK 94
E L + D + F LS+ PK+ + P ++ E++K
Sbjct: 249 NLELPLLSDYDTDNVTAKFEGAKLSIKFGELSQTKPKETLITPPSQKIEQQK 404
>CF806552
Length = 508
Score = 33.1 bits (74), Expect = 0.076
Identities = 23/63 (36%), Positives = 29/63 (45%)
Frame = +3
Query: 37 GERPFDGTKWKRFKKEFELPKYCNEDAIRGNFMQNILSVVLPKKVDLIPQEEQEEEEKIP 96
GER + K F+ F LP + D I GNF IL V +PK+ QE E
Sbjct: 306 GERQTNEQKRIHFELAFPLPPDSDVDNISGNFDSEILHVHVPKRA-----SHQEHRESGH 470
Query: 97 ELE 99
E+E
Sbjct: 471 EIE 479
>TC227161 similar to UP|Q9LUM0 (Q9LUM0) Emb|CAB36798.1, partial (23%)
Length = 1726
Score = 31.2 bits (69), Expect = 0.29
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 2/57 (3%)
Frame = -1
Query: 60 NEDAIRGNFMQNILSVVLPKKV--DLIPQEEQEEEEKIPELEDLDKYQEKNTYKSLG 114
N D +R M+NI+S + PK E + K+PE E KY E N++ SLG
Sbjct: 427 NIDELRDPDMENIISSIDPKLF*ESKAVSSTDENDCKVPESE---KYDEVNSFPSLG 266
>TC230087 similar to GB|BAB08313.1|9757977|AB018107 heat shock hsp20
protein-like {Arabidopsis thaliana;} , partial (75%)
Length = 571
Score = 30.0 bits (66), Expect = 0.64
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 3/63 (4%)
Frame = +3
Query: 20 WRRE---QDRDTIELHLPGNGERPFDGTKWKRFKKEFELPKYCNEDAIRGNFMQNILSVV 76
WR E +++DT+ H+ ER GT F +E ELP+ D I+ +L++V
Sbjct: 270 WREEPQAKEKDTV-WHV---AER---GTGKGGFSREIELPENVKVDQIKAQVENGVLTIV 428
Query: 77 LPK 79
+PK
Sbjct: 429 VPK 437
>TC213861
Length = 518
Score = 28.1 bits (61), Expect = 2.4
Identities = 16/62 (25%), Positives = 32/62 (50%)
Frame = +2
Query: 79 KKVDLIPQEEQEEEEKIPELEDLDKYQEKNTYKSLGFGGRDREEEIGTLSEYTYRTDNKF 138
KK D ++E E I + K++Y SL +++ EE+G ++ T + ++++
Sbjct: 323 KKNDFDQKKEVTGESNI-------SLRPKSSYMSLDSDEKNKHEEVGAVASTTSQNNDEY 481
Query: 139 GE 140
GE
Sbjct: 482 GE 487
>CA936501
Length = 266
Score = 28.1 bits (61), Expect = 2.4
Identities = 12/37 (32%), Positives = 25/37 (67%)
Frame = +1
Query: 79 KKVDLIPQEEQEEEEKIPELEDLDKYQEKNTYKSLGF 115
+K ++IP+EE+EEEE++ + L +Q +++ + F
Sbjct: 52 RKKEVIPEEEEEEEERLQQ*IKLILFQP*SSHNEICF 162
>TC223164
Length = 686
Score = 26.9 bits (58), Expect = 5.5
Identities = 20/63 (31%), Positives = 31/63 (48%), Gaps = 6/63 (9%)
Frame = +3
Query: 57 KYCNEDAIRGNFMQNILSVVLPKKVDLIPQEEQEEEEKIP------ELEDLDKYQEKNTY 110
K+C + F I + +L K + LIPQ+E E E+ +P E +D + EK
Sbjct: 294 KFCANEKPTVRFKIGIKNKLLGKSIGLIPQKE-EHEKTLPWFAMEDEKDDRKRRAEKILR 470
Query: 111 KSL 113
+SL
Sbjct: 471 ESL 479
>TC231862 similar to GB|AAL60196.1|18139887|AF441079 O-linked N-acetyl
glucosamine transferase {Arabidopsis thaliana;} ,
partial (25%)
Length = 750
Score = 26.9 bits (58), Expect = 5.5
Identities = 13/28 (46%), Positives = 15/28 (53%)
Frame = -1
Query: 56 PKYCNEDAIRGNFMQNILSVVLPKKVDL 83
P YC I G+ Q + SV LP VDL
Sbjct: 102 PSYCKLPKIWGHMQQLVYSVELPYPVDL 19
>TC207950 similar to GB|AAT06459.1|46931310|BT012640 At3g57910 {Arabidopsis
thaliana;} , partial (72%)
Length = 1259
Score = 26.6 bits (57), Expect = 7.1
Identities = 14/27 (51%), Positives = 20/27 (73%)
Frame = +1
Query: 75 VVLPKKVDLIPQEEQEEEEKIPELEDL 101
+V P+K D + + E+EEEE+I E EDL
Sbjct: 589 IVEPQKNDDVAEGEEEEEEEITE-EDL 666
>TC207951 similar to GB|AAT06459.1|46931310|BT012640 At3g57910 {Arabidopsis
thaliana;} , partial (23%)
Length = 707
Score = 26.6 bits (57), Expect = 7.1
Identities = 14/27 (51%), Positives = 20/27 (73%)
Frame = +3
Query: 75 VVLPKKVDLIPQEEQEEEEKIPELEDL 101
+V P+K D + + E+EEEE+I E EDL
Sbjct: 45 IVEPQKNDDVAEGEEEEEEEITE-EDL 122
>TC233709
Length = 754
Score = 26.2 bits (56), Expect = 9.3
Identities = 9/20 (45%), Positives = 14/20 (70%)
Frame = -2
Query: 41 FDGTKWKRFKKEFELPKYCN 60
++GTKW+ K EF L + C+
Sbjct: 165 YEGTKWQNPKSEFALKQTCS 106
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,111,096
Number of Sequences: 63676
Number of extensions: 88599
Number of successful extensions: 495
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 487
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 491
length of query: 184
length of database: 12,639,632
effective HSP length: 91
effective length of query: 93
effective length of database: 6,845,116
effective search space: 636595788
effective search space used: 636595788
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC126009.2


