

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125480.15 - phase: 0
(362 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
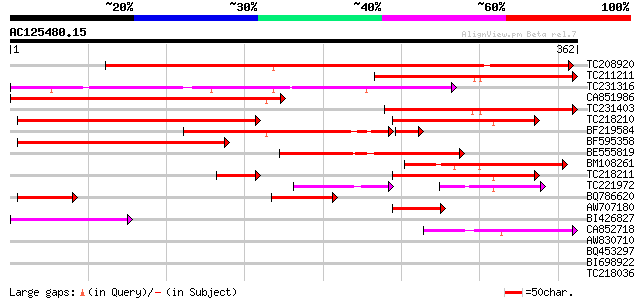
Score E
Sequences producing significant alignments: (bits) Value
TC208920 weakly similar to GB|AAT42380.1|48427662|BT014888 At4g3... 231 3e-61
TC211211 similar to UP|Q9SPV4 (Q9SPV4) S-adenosyl-L-methionine:s... 180 8e-46
TC231316 similar to UP|Q9FLN8 (Q9FLN8) S-adenosyl-L-methionine:s... 178 4e-45
CA851986 weakly similar to GP|13365694|dbj caffeine synthase {Co... 176 1e-44
TC231403 weakly similar to UP|Q9SPV4 (Q9SPV4) S-adenosyl-L-methi... 171 4e-43
TC218210 weakly similar to UP|Q84PP8 (Q84PP8) 3,7-dimethylxanthi... 164 6e-41
BF219584 weakly similar to GP|20271026|gb| N-methyltransferase {... 120 2e-30
BF595358 weakly similar to GP|13366161|dbj S-adenosyl-L-methioni... 127 8e-30
BE555819 109 2e-24
BM108261 weakly similar to GP|6002712|gb|A S-adenosyl-L-methioni... 87 1e-17
TC218211 weakly similar to UP|Q84PP8 (Q84PP8) 3,7-dimethylxanthi... 71 9e-13
TC221972 weakly similar to UP|Q8W414 (Q8W414) S-adenosyl-L-methi... 47 2e-12
BQ786620 weakly similar to GP|13235641|emb S-adenosyl-L-methioni... 55 4e-08
AW707180 55 5e-08
BI426827 weakly similar to GP|9758122|dbj| S-adenosyl-L-methioni... 54 1e-07
CA852718 46 2e-05
AW830710 36 0.024
BQ453297 33 0.15
BI698922 30 1.7
TC218036 similar to UP|Q9SEZ9 (Q9SEZ9) At2g40160/T7M7.25 (At2g40... 30 1.7
>TC208920 weakly similar to GB|AAT42380.1|48427662|BT014888 At4g36470
{Arabidopsis thaliana;} , partial (48%)
Length = 1028
Score = 231 bits (590), Expect = 3e-61
Identities = 117/305 (38%), Positives = 188/305 (61%), Gaps = 6/305 (1%)
Frame = +2
Query: 62 GSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMG 121
G NTL +I DI + ++ +++ H S E+ +Y NDLP NDFN+IF +L F++ L +
Sbjct: 2 GPNTLSIIKDIFQAIQGTSQRIMHHSTEFRVYFNDLPTNDFNSIFKALPEFQKLLRQDRK 181
Query: 122 TEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN------NKGNIYIS 175
F G PGSF+GR+FP LHFVHSS+SLHWLS+VP + NKG +YI
Sbjct: 182 NGFPSIFMGGYPGSFYGRLFPNSYLHFVHSSFSLHWLSRVPPSLYDEHKRPLNKGCVYIC 361
Query: 176 STSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELL 235
+SP V +AYY+QFQ DFSLFL+ R+EE+V GG M++ F+GR+ + + WE+L
Sbjct: 362 ESSPEVVSQAYYQQFQEDFSLFLRSRSEELVVGGRMVLIFLGRRGPEHVDRGNSFFWEIL 541
Query: 236 AMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARDD 295
+ + +V +G +++EK +++++ Y PS E++ EV EGS + +LE+ E++ + +
Sbjct: 542 SRSFAILVSQGEVEQEKFDSYDVHFYAPSREEIEEEVRKEGSLKLERLEMFEMDKSHNEH 721
Query: 296 FESESFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINIT 355
ES+G VA +RA+ E ++ HFGEG+++ +F + K + + +K + I+
Sbjct: 722 GSDESYGT---QVAVAVRAIQESMISHHFGEGILESLFQNFAKLVDEEMAKEDIRPISFV 892
Query: 356 ILLAK 360
++L K
Sbjct: 893 LVLRK 907
>TC211211 similar to UP|Q9SPV4 (Q9SPV4) S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase, partial (28%)
Length = 665
Score = 180 bits (457), Expect = 8e-46
Identities = 90/137 (65%), Positives = 113/137 (81%), Gaps = 8/137 (5%)
Frame = +1
Query: 234 LLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNAR 293
L A ALNDMVL+GIIKEE+L+TFNIP Y PSPSEV+LEV+ EGSF N+LE+SEVNWNA
Sbjct: 34 LWATALNDMVLQGIIKEEQLDTFNIPQYTPSPSEVELEVLKEGSFASNRLEVSEVNWNAF 213
Query: 294 DD-----FESE---SFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNS 345
DD FESE + D GYNVAQCMRA+AEP+LVSHFGE +++E+F+RY++ LTDR S
Sbjct: 214 DDWNTLEFESERSDTLSDGGYNVAQCMRAVAEPMLVSHFGEAIIEEVFSRYQQILTDRMS 393
Query: 346 KGRTKFINITILLAKRS 362
K +TKFIN+T+LL +++
Sbjct: 394 KEQTKFINVTVLLTRKA 444
>TC231316 similar to UP|Q9FLN8 (Q9FLN8) S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase-like protein, partial
(76%)
Length = 1111
Score = 178 bits (451), Expect = 4e-45
Identities = 113/306 (36%), Positives = 163/306 (52%), Gaps = 21/306 (6%)
Frame = +1
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQ----RKVIYLTKPLRDEAITSMYNNTLSKSLTIAD 56
ME+ K+L M GG GE SYANNS Q R +++L + D + +AD
Sbjct: 130 MELEKLLSMKGGKGEASYANNSQAQAIHARSMLHLLRETLDRVEVVEGREV---AFVVAD 300
Query: 57 LGCSSGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKL 116
LGCS GSN++ V+ +IK + K L + PE+ + +DLP NDFNT+F L
Sbjct: 301 LGCSCGSNSINVVDVMIKHMMKRYEALGWQPPEFSAFFSDLPSNDFNTLFQLLPP----- 465
Query: 117 LDEMGTEMGPC---------FFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVPEGADN 167
L G M C F +GVPGSF+ R+FP +S+ HS++SLHWLS+VPE ++
Sbjct: 466 LANYGVSMEECLAANNHRSYFAAGVPGSFYRRLFPARSVDVFHSAFSLHWLSQVPESVED 645
Query: 168 ------NKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSD 221
NKG ++I + AY KQFQ D + FL+ R+ E+ G M + + R S
Sbjct: 646 KRSSAYNKGKVFIHGAGE-STANAYKKQFQTDLAGFLRARSVEMKREGSMFLVCLARTSV 822
Query: 222 NPTSK--ECCYIWELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFV 279
+PT + A +D+V EG+I +EK + FNIP+Y S + K V GSF
Sbjct: 823 DPTDQGGAGLLFGTHFQDAWDDLVQEGLISQEKRDNFNIPVYAASLQDFKEVVEANGSFA 1002
Query: 280 MNQLEI 285
+++LE+
Sbjct: 1003IDKLEV 1020
>CA851986 weakly similar to GP|13365694|dbj caffeine synthase {Coffea
arabica}, partial (19%)
Length = 635
Score = 176 bits (446), Expect = 1e-44
Identities = 89/182 (48%), Positives = 118/182 (63%), Gaps = 6/182 (3%)
Frame = +1
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
ME +LHMN G GE SYANNS++QRK++ K + +E IT Y+N + +ADLGCS
Sbjct: 88 MESKLLLHMNSGKGERSYANNSMLQRKLMIKGKHILEETITRFYSNYSPSCMKVADLGCS 267
Query: 61 SGSNTLLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEM 120
G NTLLVI +II +V+ C +LN + P + YLNDL GNDFNT F SL F ++L ++
Sbjct: 268 VGPNTLLVISNIIDIVDTTCTRLNQEPPTFQFYLNDLFGNDFNTTFKSLPDFYKRLDEDK 447
Query: 121 GTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP------EGADNNKGNIYI 174
G + G CF + PGSFHGR+FP S++ HS+ SLHWLS+ P E NKG+ +I
Sbjct: 448 GHKFGSCFINATPGSFHGRLFPNNSINLFHSANSLHWLSQDPLLEFTKEAESFNKGHCHI 627
Query: 175 SS 176
S
Sbjct: 628 VS 633
>TC231403 weakly similar to UP|Q9SPV4 (Q9SPV4)
S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase, partial (29%)
Length = 642
Score = 171 bits (434), Expect = 4e-43
Identities = 84/131 (64%), Positives = 108/131 (82%), Gaps = 8/131 (6%)
Frame = +2
Query: 240 NDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNWNARD----- 294
+DMVL+GII+EE+L+TFNIP Y PSPSEVKLEV+ EGSF +N+LE+SEVNWNA D
Sbjct: 2 SDMVLQGIIREEQLDTFNIPQYTPSPSEVKLEVLKEGSFAINRLEVSEVNWNALDEWNAL 181
Query: 295 DFESE---SFGDDGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKF 351
DFESE S D GYNVAQCMRA+AEP+L+SHFGE +++E+F RY++ L +R SK +TKF
Sbjct: 182 DFESERSESLSDGGYNVAQCMRAVAEPMLISHFGEAIIEEVFCRYQQILAERMSKEKTKF 361
Query: 352 INITILLAKRS 362
IN+TILL +++
Sbjct: 362 INVTILLTRKA 394
>TC218210 weakly similar to UP|Q84PP8 (Q84PP8) 3,7-dimethylxanthine
N-methyltransferase, partial (27%)
Length = 845
Score = 164 bits (415), Expect = 6e-41
Identities = 76/155 (49%), Positives = 104/155 (67%)
Frame = +1
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNT 65
+LHMNGG GE SY NN L+Q+K++ KP+ +E I +Y + + + +LGCS G N
Sbjct: 19 LLHMNGGKGERSYTNNCLLQKKLMLKAKPILEETIMRLYRDFSPNCMKVTNLGCSVGPNA 198
Query: 66 LLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMG 125
LLVI +II +V C LN + P++ YLNDL GN FNTIF SL F +L+++ G + G
Sbjct: 199 LLVISNIIDIVNTACTSLNREPPKFQFYLNDLFGNGFNTIFKSLPNFYTRLVEDKGHKFG 378
Query: 126 PCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSK 160
PCF + PGSF+GR+FP S++ HSS SLHWLS+
Sbjct: 379 PCFVNATPGSFYGRLFPSNSINLFHSSNSLHWLSQ 483
Score = 69.7 bits (169), Expect = 2e-12
Identities = 38/98 (38%), Positives = 60/98 (60%), Gaps = 4/98 (4%)
Frame = +1
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW-NARDDFESESFGD 303
+G+I+EEKL++FNIP+Y P+ E++ + EGSF + + EI + W ++ SF D
Sbjct: 481 QGLIEEEKLDSFNIPVYEPTVEEIRHVIEEEGSFFVQRFEILTLPWVEGLNEGGDNSFLD 660
Query: 304 DGYN---VAQCMRALAEPLLVSHFGEGVVKEIFNRYKK 338
+A+ +RA EPLL + FGE V+ E+F Y+K
Sbjct: 661 GNIKAR*MAKHVRAAMEPLLSTKFGEEVINEVFIMYQK 774
>BF219584 weakly similar to GP|20271026|gb| N-methyltransferase {Coffea
canephora}, partial (18%)
Length = 597
Score = 120 bits (302), Expect(2) = 2e-30
Identities = 65/140 (46%), Positives = 88/140 (62%), Gaps = 6/140 (4%)
Frame = +3
Query: 112 FKEKLLDEMGTEMGPCFFSGVPGSFHGRIFPLQSLHFVHSSYSLHWLSKVP------EGA 165
F +L+++ G + GPCF + PGS++GR+FP ++F HSS SLHWLS+ P E
Sbjct: 36 FYTRLVEDKGHKFGPCFINATPGSYYGRLFPSNFINFFHSSTSLHWLSQDPLLGLTREAE 215
Query: 166 DNNKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTS 225
N GN YI STSP V KAY KQ Q F LFLK R EE++ GG M++ F GR N T
Sbjct: 216 SLN*GNCYIVSTSPPEVYKAYLKQSQEGFKLFLKSRWEELMPGGAMVLVFPGR---NETP 386
Query: 226 KECCYIWELLAMALNDMVLE 245
+ + E++++ LNDM+LE
Sbjct: 387 RRS--LREVISLTLNDMLLE 440
Score = 29.3 bits (64), Expect(2) = 2e-30
Identities = 10/18 (55%), Positives = 17/18 (93%)
Frame = +2
Query: 247 IIKEEKLNTFNIPIYYPS 264
+I++EKL++FNIP+Y P+
Sbjct: 542 LIEDEKLDSFNIPVYEPT 595
>BF595358 weakly similar to GP|13366161|dbj S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase {Atropa belladonna},
partial (14%)
Length = 415
Score = 127 bits (319), Expect = 8e-30
Identities = 61/135 (45%), Positives = 91/135 (67%)
Frame = +2
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCSSGSNT 65
+LHMNGG GE SYA++S +Q+K++ P+ +E I +Y+++ + +ADLGCS G N+
Sbjct: 11 LLHMNGGKGERSYAHHSSLQKKLMLKATPILEETIMKLYHDSSPSCMKVADLGCSVGPNS 190
Query: 66 LLVILDIIKVVEKLCRKLNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLLDEMGTEMG 125
LLVI +II +V+ C LN + P + YLNDL GNDFNTIF SL +L ++ G ++G
Sbjct: 191 LLVISNIINIVDTTCTILNCEPPTFPFYLNDLLGNDFNTIFQSLPDSHTRLGED*GHKLG 370
Query: 126 PCFFSGVPGSFHGRI 140
CF + PGS +G++
Sbjct: 371 LCFINATPGSCYGKV 415
>BE555819
Length = 425
Score = 109 bits (272), Expect = 2e-24
Identities = 56/118 (47%), Positives = 79/118 (66%)
Frame = +2
Query: 173 YISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIW 232
+I STSP + KAY KQFQ DF LFLK R+EE+V GG M++ +G + P C
Sbjct: 23 HIVSTSPPEIYKAYVKQFQQDFKLFLKSRSEELVPGGAMVLVVLGNH-ETPRRIGC---- 187
Query: 233 ELLAMALNDMVLEGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW 290
EL+++ LNDM LEG+I+EEKL++FNIP+Y P+ E++ + EGSF + + EI + W
Sbjct: 188 ELVSLKLNDMFLEGLIEEEKLDSFNIPVYEPTVEEIRHVIEEEGSFFVQRFEILTLPW 361
>BM108261 weakly similar to GP|6002712|gb|A S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase {Clarkia breweri},
partial (12%)
Length = 717
Score = 87.0 bits (214), Expect = 1e-17
Identities = 52/116 (44%), Positives = 72/116 (61%), Gaps = 12/116 (10%)
Frame = -1
Query: 253 LNTF-NIPIYYPSPSEVKLEVITEGSFVMNQ----LEISEVNWNARDDFES------ESF 301
L TF IP Y S + E+ G F+ +E+SEVNWNA D++ + + F
Sbjct: 552 LGTFLTIPFIYHSXPHFESEI---GKFLKXDNLPLIEVSEVNWNAHDEWNAP*VWILKYF 382
Query: 302 GD-DGYNVAQCMRALAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITI 356
G+ D NV QCMRA+AE +LVSHFGE + E+F+RY++ L+DR SK +TKF +TI
Sbjct: 381 GNGDTCNVTQCMRAVAESMLVSHFGEAITDELFSRYQEILSDRMSKEKTKFTIVTI 214
>TC218211 weakly similar to UP|Q84PP8 (Q84PP8) 3,7-dimethylxanthine
N-methyltransferase, partial (17%)
Length = 645
Score = 70.9 bits (172), Expect = 9e-13
Identities = 39/98 (39%), Positives = 60/98 (60%), Gaps = 4/98 (4%)
Frame = +1
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSFVMNQLEISEVNW-NARDDFESESFGD 303
EG+I+EEKL++FNIP+Y P+ E++ + EGSF + + EI + W ++ SF D
Sbjct: 169 EGLIEEEKLDSFNIPVYEPTVEEIRHVIEEEGSFFVQRFEILTLPWVEGLNEGGDNSFLD 348
Query: 304 DGYN---VAQCMRALAEPLLVSHFGEGVVKEIFNRYKK 338
+A+ +RA EPLL + FGE V+ E+F Y+K
Sbjct: 349 GNIKAR*MAKHVRAAMEPLLSTKFGEEVINEVFIMYQK 462
Score = 45.1 bits (105), Expect = 5e-05
Identities = 18/28 (64%), Positives = 23/28 (81%)
Frame = +2
Query: 133 PGSFHGRIFPLQSLHFVHSSYSLHWLSK 160
PGSF+GR+FP S++ HSS SLHWLS+
Sbjct: 8 PGSFYGRLFPSNSINLFHSSNSLHWLSQ 91
>TC221972 weakly similar to UP|Q8W414 (Q8W414)
S-adenosyl-L-methionine:salicylic acid calboxyl
methyltransferase-like protein, partial (9%)
Length = 702
Score = 46.6 bits (109), Expect(2) = 2e-12
Identities = 27/64 (42%), Positives = 37/64 (57%)
Frame = +2
Query: 182 VVKAYYKQFQIDFSLFLKCRAEEIVEGGCMIITFVGRKSDNPTSKECCYIWELLAMALND 241
V +A QF F+LFLK RAEE++ GG M++ FVGR + + W L+ + L D
Sbjct: 5 VYQAXXXQFSPYFNLFLKSRAEELLRGGGMVLRFVGRDE----TFDIITPWGLIGLVLID 172
Query: 242 MVLE 245
MV E
Sbjct: 173 MVSE 184
Score = 43.1 bits (100), Expect(2) = 2e-12
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Frame = +1
Query: 275 EGSFVMNQLEISEVNWNARDDFESESFGDDGYN-------VAQCMRALAEPLLVSHFGEG 327
EGSF + +LE + W+ D + GD + +A+ +RA EP + + FGEG
Sbjct: 184 EGSFTLEKLETFKSRWD--DGLKENGNGDFVLDTNVRAKFIAKYVRATTEPFMTARFGEG 357
Query: 328 VVKEIFNRYKKYLTD 342
++ E+F +Y+ + +
Sbjct: 358 IIDELFPKYRNKVAE 402
>BQ786620 weakly similar to GP|13235641|emb S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase {Stephanotis
floribunda}, partial (21%)
Length = 429
Score = 55.5 bits (132), Expect = 4e-08
Identities = 26/42 (61%), Positives = 31/42 (72%)
Frame = +2
Query: 168 NKGNIYISSTSPLNVVKAYYKQFQIDFSLFLKCRAEEIVEGG 209
NKGN ++ STSP V KAY+KQFQ F FLK R+EE+V GG
Sbjct: 302 NKGNCHLVSTSPSEVYKAYFKQFQEGFKSFLKSRSEELVPGG 427
Score = 42.4 bits (98), Expect = 3e-04
Identities = 19/38 (50%), Positives = 27/38 (71%)
Frame = +3
Query: 6 VLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSM 43
+LHMNGG GE SYAN S Q+K++ K + +E IT++
Sbjct: 150 LLHMNGGKGERSYANYSSFQKKLMLKAKSILEETITTL 263
>AW707180
Length = 210
Score = 55.1 bits (131), Expect = 5e-08
Identities = 26/34 (76%), Positives = 29/34 (84%)
Frame = -2
Query: 245 EGIIKEEKLNTFNIPIYYPSPSEVKLEVITEGSF 278
+GIIKEE+L TFNIP Y PSPSEVKLEV+ EG F
Sbjct: 104 QGIIKEEQLETFNIPYYTPSPSEVKLEVLKEG*F 3
>BI426827 weakly similar to GP|9758122|dbj| S-adenosyl-L-methionine:salicylic
acid carboxyl methyltransferase-like protein, partial
(23%)
Length = 422
Score = 53.5 bits (127), Expect = 1e-07
Identities = 32/78 (41%), Positives = 42/78 (53%)
Frame = +3
Query: 1 MEVAKVLHMNGGVGETSYANNSLVQRKVIYLTKPLRDEAITSMYNNTLSKSLTIADLGCS 60
ME+ ++L M GG GE SYANNS Q L EA+ + + + DLGCS
Sbjct: 168 MELERLLSMKGGKGEGSYANNSQAQAIHAKSMHHLLKEALDGVQLQAPNMPFVVVDLGCS 347
Query: 61 SGSNTLLVILDIIKVVEK 78
GSNT+ V+ IIK + K
Sbjct: 348 CGSNTINVVDLIIKHIIK 401
>CA852718
Length = 414
Score = 46.2 bits (108), Expect = 2e-05
Identities = 33/102 (32%), Positives = 49/102 (47%), Gaps = 4/102 (3%)
Frame = +2
Query: 265 PSEVKLEVITEGSFVMNQLEISEVNWNARDDFESESFGDDGYNVAQCM----RALAEPLL 320
P ++ + EGSF + QLEI + W+ E + G D AQ M RA+ EPLL
Sbjct: 23 PGXIRHVIQEEGSFFLQQLEILILPWD-----EGLNEGVDANIKAQFMAKVARAIMEPLL 187
Query: 321 VSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAKRS 362
+ FG V+ E+F RY+K L + + I + K +
Sbjct: 188 SAKFGREVIIEVFIRYEKKLAQLMEVEKLESTTFVISMTKNA 313
>AW830710
Length = 137
Score = 36.2 bits (82), Expect = 0.024
Identities = 15/22 (68%), Positives = 18/22 (81%)
Frame = +1
Query: 7 LHMNGGVGETSYANNSLVQRKV 28
LHMNGG G+ SYANNS +QR +
Sbjct: 52 LHMNGGKGQRSYANNSSLQRTI 117
>BQ453297
Length = 430
Score = 33.5 bits (75), Expect = 0.15
Identities = 23/61 (37%), Positives = 33/61 (53%), Gaps = 10/61 (16%)
Frame = +2
Query: 312 MRALAEPLLVSHFGEGVVKEIFNRYKKYLTD----RNSKGRTKFINIT------ILLAKR 361
+RA+ EPLL + FG V+ E+F R+KK + +G T I++T LL KR
Sbjct: 8 IRAVMEPLLSTKFGGEVINELFIRFKKKIEQIMEVEKLEGATLVISMTKNH*RSYLLLKR 187
Query: 362 S 362
S
Sbjct: 188 S 190
>BI698922
Length = 310
Score = 30.0 bits (66), Expect = 1.7
Identities = 13/46 (28%), Positives = 26/46 (56%)
Frame = +3
Query: 315 LAEPLLVSHFGEGVVKEIFNRYKKYLTDRNSKGRTKFINITILLAK 360
+ EPLL + FG V+ E+F R++K + ++++ + I + K
Sbjct: 3 IMEPLLSTKFGAEVINELFTRFQKKIMQLMEVEKSEWATLMISMIK 140
>TC218036 similar to UP|Q9SEZ9 (Q9SEZ9) At2g40160/T7M7.25 (At2g40160
protein), partial (34%)
Length = 1038
Score = 30.0 bits (66), Expect = 1.7
Identities = 28/105 (26%), Positives = 44/105 (41%), Gaps = 5/105 (4%)
Frame = +1
Query: 83 LNHKSPEYMIYLNDLPGNDFNTIFTSLDIFKEKLL----DEMGTEMGPCFFSGVPGSF-H 137
+N K +Y+I FNT ++ F K+L DE TE + VP +
Sbjct: 118 MNWKDVDYLI---------FNTYIWWMNTFSMKVLRGSFDEGSTE-----YDEVPRPIAY 255
Query: 138 GRIFPLQSLHFVHSSYSLHWLSKVPEGADNNKGNIYISSTSPLNV 182
GR+ W V + D+N+ ++ SSTSPL++
Sbjct: 256 GRVLKT-------------WSKWVDDNIDSNRTKVFFSSTSPLHI 351
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,263,742
Number of Sequences: 63676
Number of extensions: 228419
Number of successful extensions: 1165
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 1146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1150
length of query: 362
length of database: 12,639,632
effective HSP length: 98
effective length of query: 264
effective length of database: 6,399,384
effective search space: 1689437376
effective search space used: 1689437376
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC125480.15


