

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.9 + phase: 0
(477 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
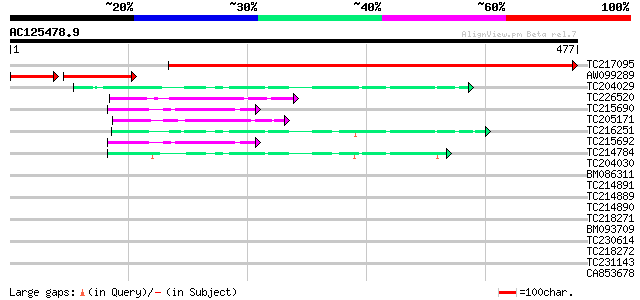
Score E
Sequences producing significant alignments: (bits) Value
TC217095 similar to PDB|1I24_A.0|33357043|1I24_A Chain A, High R... 662 0.0
AW099289 PIR|T05311|T05 sulfolipid biosynthesis protein SQD1 ch... 96 2e-31
TC204029 homologue to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydr... 48 8e-06
TC226520 homologue to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimera... 46 4e-05
TC215690 similar to UP|GAE2_CYATE (O65781) UDP-glucose 4-epimera... 45 5e-05
TC205171 similar to PIR|E86431|E86431 T5I8.7 protein - Arabidops... 44 2e-04
TC216251 homologue to UP|Q9SMJ5 (Q9SMJ5) DTDP-glucose 4-6-dehydr... 44 2e-04
TC215692 similar to UP|GAE2_CYATE (O65781) UDP-glucose 4-epimera... 43 4e-04
TC214784 similar to UP|Q9LZI2 (Q9LZI2) DTDP-glucose 4-6-dehydrat... 42 5e-04
TC204030 homologue to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydr... 41 0.001
BM086311 homologue to PIR|T00419|T00 dTDP-glucose 4-6-dehydratas... 39 0.004
TC214891 homologue to PIR|C86216|C86216 protein T23G18.6 [import... 39 0.005
TC214889 homologue to PIR|C86216|C86216 protein T23G18.6 [import... 38 0.009
TC214890 homologue to PIR|C86216|C86216 protein T23G18.6 [import... 38 0.011
TC218271 homologue to UP|Q9AV98 (Q9AV98) UDP-D-glucuronate carbo... 37 0.015
BM093709 similar to SP|O65781|GAE2 UDP-glucose 4-epimerase GEPI4... 37 0.015
TC230614 similar to UP|O24465 (O24465) Thymidine diphospho-gluco... 36 0.033
TC218272 homologue to UP|Q9SMJ5 (Q9SMJ5) DTDP-glucose 4-6-dehydr... 35 0.056
TC231143 35 0.073
CA853678 similar to SP|O65781|GAE2_ UDP-glucose 4-epimerase GEPI... 34 0.13
>TC217095 similar to PDB|1I24_A.0|33357043|1I24_A Chain A, High Resolution
Crystal Structure Of The Wild-Type Protein Sqd1, With
Nad And Udp-Glucose. {Arabidopsis thaliana;} , partial
(84%)
Length = 1346
Score = 662 bits (1709), Expect = 0.0
Identities = 317/344 (92%), Positives = 334/344 (96%)
Frame = +3
Query: 134 ISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDR 193
ISSIQ+R+QCWKSLTGK+IELYIGDIC+FEFLSETFKS+EPDAVVHFGEQRSAPYSMIDR
Sbjct: 6 ISSIQNRLQCWKSLTGKTIELYIGDICDFEFLSETFKSFEPDAVVHFGEQRSAPYSMIDR 185
Query: 194 SRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTD 253
SRAVYTQQNNV+GTLNVLFAIKE+RE CHLVKLGTMGEYGTPNIDIEEGYITITHNGRTD
Sbjct: 186 SRAVYTQQNNVIGTLNVLFAIKEFREQCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTD 365
Query: 254 TLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELCNRF 313
TLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEEL NRF
Sbjct: 366 TLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQGVVYGVRTDETAMHEELYNRF 545
Query: 314 DYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRV 373
DYD IFGTALNRFCVQAAVGHPLTVYGKGGQTR FLDIRDTVQCVELAIANPA PGEFRV
Sbjct: 546 DYDGIFGTALNRFCVQAAVGHPLTVYGKGGQTRGFLDIRDTVQCVELAIANPAKPGEFRV 725
Query: 374 FNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPNPRVELEEHYYNCKNTKLVDLGLKPH 433
FNQFTEQF V +LA LVTKAGEKLG++VKTI+VPNPRVE EEHYYNCKNTKLVDLGL+PH
Sbjct: 726 FNQFTEQFSVNQLANLVTKAGEKLGIEVKTINVPNPRVEAEEHYYNCKNTKLVDLGLRPH 905
Query: 434 FLSDSLIDSLLNFAVQYKDRVDTKQIMPGVSWRKVGVKTKTLTS 477
FLSDSL+DSLLNFAVQYKDRVDTKQIMP VSW+K+GVKTKT+T+
Sbjct: 906 FLSDSLLDSLLNFAVQYKDRVDTKQIMPSVSWKKIGVKTKTVTA 1037
>AW099289 PIR|T05311|T05 sulfolipid biosynthesis protein SQD1 chloroplast
[validated] - Arabidopsis thaliana, partial (4%)
Length = 324
Score = 95.9 bits (237), Expect(2) = 2e-31
Identities = 45/61 (73%), Positives = 50/61 (81%)
Frame = +2
Query: 46 QLFLREQKPRKSLAVVNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLS 105
+LFL EQ PRK VV+A+ +ST Q PVQTSS DPFK +RVMVIGGDGYCGWATALHLS
Sbjct: 140 RLFLPEQNPRKDSLVVHAAAVSTSQGTPVQTSSSDPFKQQRVMVIGGDGYCGWATALHLS 319
Query: 106 N 106
N
Sbjct: 320 N 322
Score = 58.5 bits (140), Expect(2) = 2e-31
Identities = 28/41 (68%), Positives = 31/41 (75%)
Frame = +3
Query: 1 MAQLLSSSCSLTFSASNKPCLKPFHQCSTSFSNTVVCDNSK 41
MAQLLSSSCSLT + NKP LKPF +CST FS V C+N K
Sbjct: 3 MAQLLSSSCSLTVCSRNKPYLKPFDKCSTPFSMKVACNNCK 125
>TC204029 homologue to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (92%)
Length = 1640
Score = 48.1 bits (113), Expect = 8e-06
Identities = 81/338 (23%), Positives = 132/338 (38%), Gaps = 1/338 (0%)
Frame = +2
Query: 54 PRKSLAVVNASTISTGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEVAI 113
PR LA + + TG+ PV G + +R++V GG G+ G L +G +V +
Sbjct: 329 PRTGLARFSGTRPRTGR-VPV----GIGGRRQRIVVTGGAGFVGSHLVDKLIARGDDVIV 493
Query: 114 VDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKSLTGKSIELYIGDICEFEFLSETFKSYE 173
+DN ++ + L EL D+ E L E
Sbjct: 494 IDNFFTGRKENLVHL------------------FGNPRFELIRHDVVEPILL-------E 598
Query: 174 PDAVVHFGEQRSAPYSMID-RSRAVYTQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEY 232
D + H + P S + + V T + NV+GTLN+L K R + T Y
Sbjct: 599 VDQIYHL----ACPASPVHYKYNPVKTIKTNVMGTLNMLGLAK--RIGARFLLTSTSEVY 760
Query: 233 GTPNIDIEEGYITITHNGRTDTLPYPKQASSFYHLSKVHDSHNIAFTCKAWGIRATDLNQ 292
G P L +P++ + + +++ + + + A D ++
Sbjct: 761 GDP-------------------LEHPQKETYWGNVNPIGERSCYDEGKRTAETLAMDYHR 883
Query: 293 GVVYGVRTDETAMHEELCNRFDYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIR 352
G GV + R D G ++ F QA PLTVYG G QTR+F +
Sbjct: 884 GA--GVEVRIARIFNTYGPRMCLDD--GRVVSNFVAQAIRKQPLTVYGDGKQTRSFQYVS 1051
Query: 353 DTVQCVELAIANPANPGEFRVFNQFTEQFKVTELAELV 390
D V + +A+ + G F + N +F + ELA++V
Sbjct: 1052DLVNGL-VALMESEHVGPFNLGN--PGEFTMLELAQVV 1156
>TC226520 homologue to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase
(Galactowaldenase) (UDP-galactose 4-epimerase) ,
complete
Length = 1290
Score = 45.8 bits (107), Expect = 4e-05
Identities = 43/162 (26%), Positives = 74/162 (45%), Gaps = 3/162 (1%)
Frame = +2
Query: 85 KRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRI-QC 143
+ ++V GG G+ G T + L G+ V+I+DN FD+ + DR+ Q
Sbjct: 50 QHILVTGGAGFIGTHTVVQLLKAGFSVSIIDN-----FDNS--------VMEAVDRVRQV 190
Query: 144 WKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNN 203
L ++++ GD+ + L + F DAV+HF ++ S+ R N
Sbjct: 191 VGPLLSQNLQFTQGDLRNRDDLEKLFSKTTFDAVIHFAGLKAVAESVAKPRRYF---DFN 361
Query: 204 VVGTLNVLFAIKEYREDC-HLVKLGTMGEYGTP-NIDIEEGY 243
+VGT+N+ + +Y +C +V + YG P I EE +
Sbjct: 362 LVGTINLYEFMAKY--NCKKMVFSSSATVYGQPEKIPCEEDF 481
>TC215690 similar to UP|GAE2_CYATE (O65781) UDP-glucose 4-epimerase GEPI48
(Galactowaldenase) (UDP-galactose 4-epimerase) , partial
(36%)
Length = 539
Score = 45.4 bits (106), Expect = 5e-05
Identities = 35/129 (27%), Positives = 60/129 (46%)
Frame = +1
Query: 83 KPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQ 142
+ K V+V GG GY G T L L G +VDNL D+ + +S R++
Sbjct: 148 RDKTVLVTGGAGYIGTHTVLQLLLGGCRTVVVDNL-----------DNSSEVSI--HRVR 288
Query: 143 CWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQN 202
G ++ + D+ + + L + F S + DAV+HF ++ S+ + + N
Sbjct: 289 ELAGEFGNNLSFHKVDLRDRDALEQIFVSTQFDAVIHFAGLKAVGESV---QKPLLYYNN 459
Query: 203 NVVGTLNVL 211
N+ GT+ +L
Sbjct: 460 NLTGTITLL 486
>TC205171 similar to PIR|E86431|E86431 T5I8.7 protein - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (97%)
Length = 1690
Score = 43.5 bits (101), Expect = 2e-04
Identities = 41/149 (27%), Positives = 63/149 (41%)
Frame = +2
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V GG GY G L L + Y V IVDNL R +L + +QD
Sbjct: 452 VLVTGGAGYIGSHATLRLLRENYRVTIVDNLSR---------GNLGAVRVLQDLFP---- 592
Query: 147 LTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNNVVG 206
++ D+ + E +++ F + DAV+HF S +D + + +N
Sbjct: 593 -EPGRLQFIYADLGDKESVNKIFSENKFDAVMHFAAVAYVGESTLDPLKYYHNITSN--- 760
Query: 207 TLNVLFAIKEYREDCHLVKLGTMGEYGTP 235
TL VL ++ +Y L+ T YG P
Sbjct: 761 TLLVLESMAKYGVKT-LIYSSTCATYGEP 844
>TC216251 homologue to UP|Q9SMJ5 (Q9SMJ5) DTDP-glucose 4-6-dehydratase ,
partial (98%)
Length = 1378
Score = 43.5 bits (101), Expect = 2e-04
Identities = 77/324 (23%), Positives = 126/324 (38%), Gaps = 5/324 (1%)
Frame = +1
Query: 86 RVMVIGGDGYCGWATALHL-SNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCW 144
R++V GG G+ G L N+ EV + DN + +D ++ W
Sbjct: 172 RILVTGGAGFIGSHLVDRLMENEKNEVIVADNY----------------FTGSKDNLKKW 303
Query: 145 KSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVYTQQNN 203
+ EL D+ E + E D + H + P S I + V T + N
Sbjct: 304 --IGHPRFELIRHDVTEPLLI-------EVDQIYHL----ACPASPIFYKYNPVKTIKTN 444
Query: 204 VVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYPKQASS 263
V+GTLN+L K R ++ T YG P L +P+ S
Sbjct: 445 VIGTLNMLGLAK--RVGARILLTSTSEVYGDP-------------------LVHPQPESY 561
Query: 264 FYHLSKVHDSHNIAFTCKAWGIRATD---LNQGVVYGVRTDETAMHEELCNRFDYDAIFG 320
+ +++ + +C G R + + +G+ + R + D G
Sbjct: 562 WGNVNPIG-----VRSCYDEGKRVAETLMFDYHRQHGIEIRIARIFNTYGPRMNIDD--G 720
Query: 321 TALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRVFNQFTEQ 380
++ F QA G PLTV G QTR+F + D V + + + +N G + N +
Sbjct: 721 RVVSNFIAQALRGEPLTVQCPGTQTRSFCYVSDLVDGL-IRLMEGSNTGPINLGN--PGE 891
Query: 381 FKVTELAELVTKAGEKLGLDVKTI 404
F +TELAE V K G+++K +
Sbjct: 892 FTMTELAETV-KELINPGVEIKMV 960
>TC215692 similar to UP|GAE2_CYATE (O65781) UDP-glucose 4-epimerase GEPI48
(Galactowaldenase) (UDP-galactose 4-epimerase) , partial
(97%)
Length = 1518
Score = 42.7 bits (99), Expect = 4e-04
Identities = 32/129 (24%), Positives = 60/129 (45%)
Frame = +2
Query: 83 KPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQ 142
+ K V+V GG GY G T L L G+ ++DNL ++ + ++ R++
Sbjct: 182 RDKTVLVTGGAGYIGSHTVLQLLLGGFRAVVLDNL-----------ENSSEVAI--HRVR 322
Query: 143 CWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQN 202
G ++ + D+ + L + F S + DAV+HF ++ S+ + + N
Sbjct: 323 ELAGEFGNNLSFHKVDLRDRAALDQIFSSTQFDAVIHFAGLKAVGESV---QKPLLYYNN 493
Query: 203 NVVGTLNVL 211
N+ GT+ +L
Sbjct: 494 NLTGTITLL 520
>TC214784 similar to UP|Q9LZI2 (Q9LZI2) DTDP-glucose 4-6-dehydratase homolog
D18 (At3g62830/F26K9_260), partial (84%)
Length = 1652
Score = 42.4 bits (98), Expect = 5e-04
Identities = 75/305 (24%), Positives = 109/305 (35%), Gaps = 16/305 (5%)
Frame = +3
Query: 83 KPKRVMVIGGDGYCGWATALHLSNKGYEVAIVDNLV---RRLFDHQLGLDSLTPISSIQD 139
K R++V GG G+ G L +G V +VDN + H G
Sbjct: 369 KGLRIVVTGGAGFVGSHLVDRLIARGDSVIVVDNFFTGRKENVMHHFG------------ 512
Query: 140 RIQCWKSLTGKSIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMID-RSRAVY 198
EL D+ E L E D + H + P S + + V
Sbjct: 513 ---------NPRFELIRHDVVEPLLL-------EVDQIYHL----ACPASPVHYKFNPVK 632
Query: 199 TQQNNVVGTLNVLFAIKEYREDCHLVKLGTMGEYGTPNIDIEEGYITITHNGRTDTLPYP 258
T + NVVGTLN+L K R + T YG P L +P
Sbjct: 633 TIKTNVVGTLNMLGLAK--RVGARFLLTSTSEVYGDP-------------------LQHP 749
Query: 259 KQASSFYHLSKVHDSHNIAFTCKAWGIRAT-----DLNQGVVYGVRTDETAMHEELCNRF 313
++ + + +++ + +C G R D ++G GV + R
Sbjct: 750 QKETYWGNVNPIG-----VRSCYDEGKRTAETLTMDYHRGA--GVEVRIARIFNTYGPRM 908
Query: 314 DYDAIFGTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPA 366
D G ++ F QA PLTVYG G QTR+F + D V+ + + N
Sbjct: 909 CLDD--GRVVSNFVAQALRKEPLTVYGDGKQTRSFQYVSDLVEGLIRLMEGEHVGPFNLG 1082
Query: 367 NPGEF 371
NPGEF
Sbjct: 1083NPGEF 1097
>TC204030 homologue to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (36%)
Length = 778
Score = 40.8 bits (94), Expect = 0.001
Identities = 26/71 (36%), Positives = 39/71 (54%)
Frame = +1
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRVFNQFTE 379
G ++ F QA PLTVYG G QTR+F + D V + +A+ + G F + N
Sbjct: 67 GRVVSNFVAQAIRKQPLTVYGDGKQTRSFQYVSDLVNGL-VALMESEHVGPFNLGN--PG 237
Query: 380 QFKVTELAELV 390
+F + ELA++V
Sbjct: 238 EFTMLELAQVV 270
>BM086311 homologue to PIR|T00419|T00 dTDP-glucose 4-6-dehydratase homolog
At2g47650 - Arabidopsis thaliana, partial (32%)
Length = 427
Score = 39.3 bits (90), Expect = 0.004
Identities = 23/59 (38%), Positives = 30/59 (49%), Gaps = 7/59 (11%)
Frame = +2
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPANPGEF 371
G ++ F QA PLTVYG G QTR+F + D V+ + + N NPGEF
Sbjct: 95 GRVVSNFVAQALRKEPLTVYGDGKQTRSFQYVSDLVEGLMRLMEGEHVGPFNLGNPGEF 271
>TC214891 homologue to PIR|C86216|C86216 protein T23G18.6 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(66%)
Length = 1257
Score = 38.9 bits (89), Expect = 0.005
Identities = 37/126 (29%), Positives = 57/126 (44%), Gaps = 12/126 (9%)
Frame = +3
Query: 333 GHPLTVYGKGGQTRAFLDIRDTVQCVELAIANP--ANPGEFRVFNQFTEQFKVTELAELV 390
G PL + G R F+ I+D ++ V L I NP AN F V N + V +LAE++
Sbjct: 345 GEPLKLVDGGQSQRTFVYIKDAIEAVLLMIENPARANGHIFNVGNP-NNEVTVRQLAEIM 521
Query: 391 TKAGEKLGLDVKTISVPNPRVELEEHY---YNCKNTKLVD-------LGLKPHFLSDSLI 440
K K+ + +T P V +E Y Y+ + ++ D LG P L+
Sbjct: 522 IKVYSKVSGE-QTPETPTVDVSSKEFYGEGYDDSDKRIPDMTIINRQLGWNPKTSLWDLL 698
Query: 441 DSLLNF 446
+S L +
Sbjct: 699 ESTLTY 716
>TC214889 homologue to PIR|C86216|C86216 protein T23G18.6 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(97%)
Length = 1671
Score = 38.1 bits (87), Expect = 0.009
Identities = 36/126 (28%), Positives = 57/126 (44%), Gaps = 12/126 (9%)
Frame = +3
Query: 333 GHPLTVYGKGGQTRAFLDIRDTVQCVELAIANP--ANPGEFRVFNQFTEQFKVTELAELV 390
G PL + G R F+ I+D ++ V L I NP AN F V N + V +LAE++
Sbjct: 798 GEPLKLVDGGQSQRTFIYIKDAIEAVLLMIENPARANGHIFNVGNP-NNEVTVRQLAEIM 974
Query: 391 TKAGEKLGLDVKTISVPNPRVELEEHY---YNCKNTKLVD-------LGLKPHFLSDSLI 440
+ K+ + +T P V +E Y Y+ + ++ D LG P L+
Sbjct: 975 IQVYSKVSGE-QTAETPTIDVSSKEFYGEGYDDSDKRIPDMTIINKQLGWNPKTSLWDLL 1151
Query: 441 DSLLNF 446
+S L +
Sbjct: 1152ESTLTY 1169
>TC214890 homologue to PIR|C86216|C86216 protein T23G18.6 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(98%)
Length = 1567
Score = 37.7 bits (86), Expect = 0.011
Identities = 36/126 (28%), Positives = 57/126 (44%), Gaps = 12/126 (9%)
Frame = +3
Query: 333 GHPLTVYGKGGQTRAFLDIRDTVQCVELAIANP--ANPGEFRVFNQFTEQFKVTELAELV 390
G PL + G R F+ I+D ++ V L I NP AN F V N + V +LAE++
Sbjct: 753 GEPLKLVDGGQSQRTFVYIKDAIEAVLLMIENPARANGHIFNVGNP-NNEVTVRQLAEMM 929
Query: 391 TKAGEKLGLDVKTISVPNPRVELEEHY---YNCKNTKLVD-------LGLKPHFLSDSLI 440
T+ K+ + + P V +E Y Y+ + ++ D LG P L+
Sbjct: 930 TQVYSKVSGEA-PLEKPTIDVSSKEFYGEGYDDSDKRIPDMTIINRQLGWNPKTSLWDLL 1106
Query: 441 DSLLNF 446
+S L +
Sbjct: 1107ESTLTY 1124
>TC218271 homologue to UP|Q9AV98 (Q9AV98) UDP-D-glucuronate carboxy-lyase ,
partial (51%)
Length = 731
Score = 37.4 bits (85), Expect = 0.015
Identities = 32/96 (33%), Positives = 41/96 (42%), Gaps = 7/96 (7%)
Frame = +1
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPANPGEFR 372
G ++ F QA G PLTV G QTR+F + D V + + N NPGEF
Sbjct: 151 GRVVSNFIAQAIRGEPLTVQSPGTQTRSFCYVSDLVDGLIRLMEGSDTGPINLGNPGEFT 330
Query: 373 VFNQFTEQFKVTELAELVTKAGEKLGLDVKTISVPN 408
+ ELAE V E + DV+ V N
Sbjct: 331 ML----------ELAETVK---ELINPDVEIKVVEN 399
>BM093709 similar to SP|O65781|GAE2 UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (32%)
Length = 385
Score = 37.4 bits (85), Expect = 0.015
Identities = 37/123 (30%), Positives = 57/123 (46%), Gaps = 3/123 (2%)
Frame = +3
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
V+V GG GY G T L L GY V VDN FD+ S T I+ + K
Sbjct: 72 VLVTGGAGYIGSHTVLQLLLSGYHVFAVDN-----FDN----SSETAINRV-------KE 203
Query: 147 LTGK---SIELYIGDICEFEFLSETFKSYEPDAVVHFGEQRSAPYSMIDRSRAVYTQQNN 203
L G+ ++ D+ + L + F + + DAV+HF ++ S +D+ + NN
Sbjct: 204 LAGEFANNLSFSKLDLRDRAALEKIFSTNKFDAVIHFAGLKAVGES-VDKPLLYF--DNN 374
Query: 204 VVG 206
++G
Sbjct: 375 LIG 383
>TC230614 similar to UP|O24465 (O24465) Thymidine diphospho-glucose
4-6-dehydratase homolog (Fragment), partial (75%)
Length = 886
Score = 36.2 bits (82), Expect = 0.033
Identities = 25/71 (35%), Positives = 35/71 (49%)
Frame = +1
Query: 320 GTALNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCVELAIANPANPGEFRVFNQFTE 379
G ++ F QA G PLTV G QTR+F + D V + + + N G + N
Sbjct: 211 GRVVSNFIAQAIRGEPLTVQVPGTQTRSFCYVSDMVDGL-IRLMEGENTGPINIGN--PG 381
Query: 380 QFKVTELAELV 390
+F + ELAE V
Sbjct: 382 EFTMIELAENV 414
>TC218272 homologue to UP|Q9SMJ5 (Q9SMJ5) DTDP-glucose 4-6-dehydratase ,
partial (48%)
Length = 650
Score = 35.4 bits (80), Expect = 0.056
Identities = 31/93 (33%), Positives = 40/93 (42%), Gaps = 7/93 (7%)
Frame = +3
Query: 323 LNRFCVQAAVGHPLTVYGKGGQTRAFLDIRDTVQCV-------ELAIANPANPGEFRVFN 375
++ F QA G PLTV G QTR+F + D V + + N NPGEF +
Sbjct: 117 VSNFIAQAIRGEPLTVQSPGTQTRSFCYVSDLVDGLIRLMEGSDTGPINLGNPGEFTML- 293
Query: 376 QFTEQFKVTELAELVTKAGEKLGLDVKTISVPN 408
ELAE V E + DV+ V N
Sbjct: 294 ---------ELAETVK---ELINPDVEIKVVEN 356
>TC231143
Length = 941
Score = 35.0 bits (79), Expect = 0.073
Identities = 20/65 (30%), Positives = 35/65 (53%), Gaps = 3/65 (4%)
Frame = -1
Query: 54 PRKSLAVVNAS-TISTGQEAPVQTSSGDPFKPKRVMV--IGGDGYCGWATALHLSNKGYE 110
P +S+ VV A +S +E+ + DP K V+V +GG G+C + + N G++
Sbjct: 389 PGRSIMVVKAEKAMSVREESGDSRGTIDPLKQHFVLVHGVGGGGWCWYKIRCLMENSGFK 210
Query: 111 VAIVD 115
V+ +D
Sbjct: 209 VSCID 195
>CA853678 similar to SP|O65781|GAE2_ UDP-glucose 4-epimerase GEPI48 (EC
5.1.3.2) (Galactowaldenase) (UDP- galactose
4-epimerase)., partial (27%)
Length = 485
Score = 34.3 bits (77), Expect = 0.13
Identities = 31/97 (31%), Positives = 45/97 (45%), Gaps = 3/97 (3%)
Frame = +3
Query: 87 VMVIGGDGYCGWATALHLSNKGYEVAIVDNLVRRLFDHQLGLDSLTPISSIQDRIQCWKS 146
++V GG GY G T L L GY V VDN FD+ S T I+ + K
Sbjct: 168 ILVTGGAGYIGSHTILQLLFGGYHVFAVDN-----FDN----SSETAINRV-------KE 299
Query: 147 LTGK---SIELYIGDICEFEFLSETFKSYEPDAVVHF 180
L G+ ++ D+ + L + F + + DAV+HF
Sbjct: 300 LAGELANNLSFCKLDLRDRAALEKIFSTVKFDAVIHF 410
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,061,576
Number of Sequences: 63676
Number of extensions: 282748
Number of successful extensions: 1491
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 1486
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1491
length of query: 477
length of database: 12,639,632
effective HSP length: 101
effective length of query: 376
effective length of database: 6,208,356
effective search space: 2334341856
effective search space used: 2334341856
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC125478.9


