

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
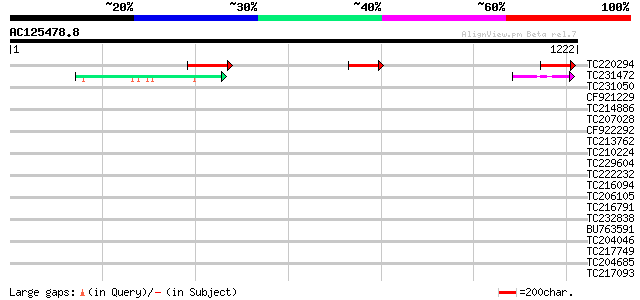
Query= AC125478.8 + phase: 0
(1222 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220294 106 7e-23
TC231472 homologue to UP|Q9FLZ6 (Q9FLZ6) Emb|CAB62594.1, partial... 51 3e-06
TC231050 weakly similar to UP|Q9SF87 (Q9SF87) F8A24.10 protein (... 37 0.053
CF921229 33 0.77
TC214886 similar to UP|Q9LNC8 (Q9LNC8) F9P14.5 protein, partial ... 33 0.77
TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%) 33 0.77
CF922292 32 1.3
TC213762 32 1.7
TC210224 similar to UP|Q84T10 (Q84T10) Unknow protein, partial (... 32 1.7
TC229604 similar to UP|Q9M276 (Q9M276) Homeobox-leucine zipper p... 32 1.7
TC222232 similar to UP|Q6UEJ2 (Q6UEJ2) Mini-chromosome maintenan... 32 1.7
TC216094 similar to UP|Q6SJQ9 (Q6SJQ9) TFIID component TAF2 (Fra... 32 2.2
TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-in... 31 3.8
TC216791 similar to UP|O23144 (O23144) Proton pump interactor (A... 30 5.0
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 30 5.0
BU763591 30 5.0
TC204046 UP|Q39835 (Q39835) Extensin, complete 30 5.0
TC217749 weakly similar to UP|Q6Z5M6 (Q6Z5M6) Transducin / WD-40... 30 6.5
TC204685 similar to UP|RK34_SPIOL (P82244) 50S ribosomal protein... 30 6.5
TC217093 30 6.5
>TC220294
Length = 1014
Score = 106 bits (264), Expect = 7e-23
Identities = 50/74 (67%), Positives = 62/74 (83%)
Frame = +2
Query: 1145 EKVRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYALRQVVSKLTPARKR 1204
+++RKFNP+EP +LPL P+ EKV LRHQ M ERK +E+WMLD ALRQVV++L PARK+
Sbjct: 563 DEMRKFNPKEPNFLPLVPEPGQEKVDLRHQMMDERKNSEDWMLDCALRQVVTQLAPARKK 742
Query: 1205 KVELLVEAFETVVP 1218
KV LLVEAFETV+P
Sbjct: 743 KVALLVEAFETVLP 784
Score = 77.4 bits (189), Expect = 4e-14
Identities = 37/74 (50%), Positives = 53/74 (71%)
Frame = +2
Query: 731 DKVRNINPRRPRELPSDANFEGEKVFLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQ 790
D++R NP+ P LP EKV L Q +ERK SE+WMLD AL++V+++LAPA+++
Sbjct: 563 DEMRKFNPKEPNFLPLVPEPGQEKVDLRHQMMDERKNSEDWMLDCALRQVVTQLAPARKK 742
Query: 791 RVTLLIEAFETLRP 804
+V LL+EAFET+ P
Sbjct: 743 KVALLVEAFETVLP 784
Score = 76.3 bits (186), Expect = 8e-14
Identities = 40/99 (40%), Positives = 62/99 (62%), Gaps = 2/99 (2%)
Frame = +2
Query: 383 EKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQ 442
+++R N + P LP EKV L Q +ERK SE+WMLD AL++V+++LAPA+++
Sbjct: 563 DEMRKFNPKEPNFLPLVPEPGQEKVDLRHQMMDERKNSEDWMLDCALRQVVTQLAPARKK 742
Query: 443 RVTLLVEAFETIRP--VQDAENGPQTSATVESHANLIQS 479
+V LLVEAFET+ P E + ++ H+ +IQ+
Sbjct: 743 KVALLVEAFETVLPAAAPKCETRVRNNSPAFGHSGIIQA 859
>TC231472 homologue to UP|Q9FLZ6 (Q9FLZ6) Emb|CAB62594.1, partial (12%)
Length = 1664
Score = 51.2 bits (121), Expect = 3e-06
Identities = 44/136 (32%), Positives = 69/136 (50%), Gaps = 2/136 (1%)
Frame = +1
Query: 1084 GGIYSEGLNQKEQKMESGNG-IVEERKEESVSKEVNKPNQKLSRNWSNLKKVVLLRRFIK 1142
G +YS+ + + K++ G +VE + E S + + ++ R +N+ V K
Sbjct: 892 GVVYSKDNDNQMIKLKFRRGKVVENQTEISSPRRLKFRRARVPREKANVNSNVR-----K 1056
Query: 1143 ALEK-VRKFNPREPRYLPLEPDSEDEKVQLRHQDMAERKGTEEWMLDYALRQVVSKLTPA 1201
+ E+ V N + P EKV LRHQDM E+K + +L+ + + SKL A
Sbjct: 1057 SFERNVACDNSNDATTGP-------EKVVLRHQDMQEKKDAQG-LLNNVIEETASKLVEA 1212
Query: 1202 RKRKVELLVEAFETVV 1217
RK KV+ LV AFETV+
Sbjct: 1213 RKSKVKALVGAFETVI 1260
Score = 48.5 bits (114), Expect = 2e-05
Identities = 74/376 (19%), Positives = 150/376 (39%), Gaps = 52/376 (13%)
Frame = +1
Query: 143 SKQVGNARKGTQKTK-----TVHSEDGNSQ-QNVKNVSMESSPFKPHDAPPSTVNECDTS 196
S +VGN K T K K T + +SQ ++K M+ S + S+V S
Sbjct: 169 SNKVGNPSKSTSKVKASSKRTTNMVKTSSQLSSLKGKEMKLSEKHVNSLNSSSVRRKQIS 348
Query: 197 TKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKHAVLSNTGKC 256
+ + + K T++E ++ + + + + + + + A +++
Sbjct: 349 SMNSSEGVGGQRYSKIKTEKEVTSSNSKAASKKLMAPSKALLTPRPSLIRVASINSRKHK 528
Query: 257 DNKVPL----------VEKEKEGGEEDNE--------GNNSYRNYSETDSDMDDEK---- 294
K+ VE E+ E E G+ + + S+ ++ DDE
Sbjct: 529 SLKIASHLKNQRSARKVEHEEHNNNEVEEKTLYVIKMGSENKTSLSDQNASYDDESYLPQ 708
Query: 295 ----KNVIELVQKAFD-------EILLPEVEDLSSEGHSKSRGNETDEVLLEKSGGKIEE 343
K+ + + K+ E E E S G+ ++ E +E L + GK ++
Sbjct: 709 LSSPKSSVSSISKSLSQEDQEESEYTTSEFEQDSFSGNHETECIENEETLAAEKKGKPKK 888
Query: 344 RNTTTFTESPKEVPKMESKQ----KSWSHLKKVILLK-RFVKALEKVRNINSRRPRQLP- 397
++ ++ K++ ++ ++ + + LK R + + N+NS +
Sbjct: 889 GGVVYSKDNDNQMIKLKFRRGKVVENQTEISSPRRLKFRRARVPREKANVNSNVRKSFER 1068
Query: 398 -------SDANFEAEKVLLNRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEA 450
+DA EKV+L Q +E KK + +L+ +++ SKL A++ +V LV A
Sbjct: 1069NVACDNSNDATTGPEKVVLRHQDMQE-KKDAQGLLNNVIEETASKLVEARKSKVKALVGA 1245
Query: 451 FETIRPVQDAENGPQT 466
FET+ + + + T
Sbjct: 1246FETVISLHEKKPSADT 1293
Score = 41.6 bits (96), Expect = 0.002
Identities = 59/223 (26%), Positives = 92/223 (40%), Gaps = 50/223 (22%)
Frame = +1
Query: 630 HNYNETDSD------MDEEKKNVIELVQKAFD-EILLPETEDLSSDDRSKSRSYGSDELL 682
HN NE + M E K + ++D E LP+ S S S+S ++
Sbjct: 592 HNNNEVEEKTLYVIKMGSENKTSLSDQNASYDDESYLPQLSSPKSSVSSISKSLSQEDQE 771
Query: 683 EK----SEGEREEMNATSFTETPKE----AKKTENKPK------SWSHLKKLIMLK-RFV 727
E SE E++ + TE + A + + KPK S + ++I LK R
Sbjct: 772 ESEYTTSEFEQDSFSGNHETECIENEETLAAEKKGKPKKGGVVYSKDNDNQMIKLKFRRG 951
Query: 728 KALDKVRNIN-PRR--------PRELPS-------------------DANFEGEKVFLNR 759
K ++ I+ PRR PRE + DA EKV L
Sbjct: 952 KVVENQTEISSPRRLKFRRARVPREKANVNSNVRKSFERNVACDNSNDATTGPEKVVLRH 1131
Query: 760 QTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETL 802
Q +E KK + +L+ +++ SKL A++ +V L+ AFET+
Sbjct: 1132QDMQE-KKDAQGLLNNVIEETASKLVEARKSKVKALVGAFETV 1257
>TC231050 weakly similar to UP|Q9SF87 (Q9SF87) F8A24.10 protein
(AT3g09850/F8A24_10), partial (12%)
Length = 1000
Score = 37.0 bits (84), Expect = 0.053
Identities = 47/189 (24%), Positives = 79/189 (40%), Gaps = 13/189 (6%)
Frame = +3
Query: 609 KDKEGREQGDAVFNGGNNSSCHNYNETDSDMDEE-KKNVIELVQKAFDEI-----LLPET 662
+D + + + ++ +++DSD+DEE ++ +E V + D I LL
Sbjct: 18 EDSSASSEPEELLGSSESNDSEYSSDSDSDIDEEVAEDYLEGVGGS-DNIMEAKWLLKPV 194
Query: 663 EDLSSDDRSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIM 722
D S DD S S Y DE LEK G + A+ +T K + S + +M
Sbjct: 195 LDESDDDSSSSSCY--DEALEKLSGFVLQ-EASREYDTKKAQSWKKRSVGSGPLALEDLM 365
Query: 723 LKRFVKALDKVRNINPRRPRELPSDA-------NFEGEKVFLNRQTSEERKKSEEWMLDY 775
L + + + + PR P+ PS A GEK L ++ +++
Sbjct: 366 LAKDPRTISARKKHVPRFPQSWPSHAQNSKASKRIHGEKKKLRKERIAVKRRERMLHRGV 545
Query: 776 ALQKVISKL 784
L+K+ SKL
Sbjct: 546 DLEKINSKL 572
>CF921229
Length = 680
Score = 33.1 bits (74), Expect = 0.77
Identities = 27/109 (24%), Positives = 49/109 (44%)
Frame = -3
Query: 263 VEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHS 322
V +EK+ E E + + + ++ +EKK V +K E+++ E +++
Sbjct: 636 VTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKKEAEVIVEEKKEVEVTEEK 457
Query: 323 KSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWSHLKK 371
K E + +K IEE+ T TE KEV ++K S +K+
Sbjct: 456 K----EVEVTEGKKEVEVIEEKKETEVTEEKKEVEVEVREEKKESEVKE 322
>TC214886 similar to UP|Q9LNC8 (Q9LNC8) F9P14.5 protein, partial (9%)
Length = 822
Score = 33.1 bits (74), Expect = 0.77
Identities = 25/98 (25%), Positives = 49/98 (49%), Gaps = 2/98 (2%)
Frame = +3
Query: 624 GNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPE--TEDLSSDDRSKSRSYGSDEL 681
G+ + +N ++ D +D+E ++ E + DE+ P +LS DD S+ ++Y + +
Sbjct: 201 GSEDTENNQDQIDLKLDDEAESDSESDVDSKDELFFPNIGMAELSEDDDSE-QTYNDESV 377
Query: 682 LEKSEGEREEMNATSFTETPKEAKKTENKPKSWSHLKK 719
E+ + E+ +A +E AK K +S +KK
Sbjct: 378 EEQPVAQHEDFSALKLSELRALAK--TRGLKGFSKMKK 485
>TC207028 similar to UP|Q9LW95 (Q9LW95) KED, partial (13%)
Length = 490
Score = 33.1 bits (74), Expect = 0.77
Identities = 27/88 (30%), Positives = 43/88 (48%), Gaps = 7/88 (7%)
Frame = +1
Query: 262 LVEKEKEGGEEDNEGNNSYR-NYSETDSDMDDEKKNVIE------LVQKAFDEILLPEVE 314
L EK K ED+EGN + + E + D EKK++ E V+ + +I ++E
Sbjct: 223 LKEKGKNDEGEDDEGNKKKKKDKKEKEKDHKKEKKDIEEGEKEDSKVEASVRDI---DIE 393
Query: 315 DLSSEGHSKSRGNETDEVLLEKSGGKIE 342
+ G K++ T + LEK GKI+
Sbjct: 394 EKKVTGKDKTKDLSTLKQKLEKINGKIQ 477
>CF922292
Length = 722
Score = 32.3 bits (72), Expect = 1.3
Identities = 26/104 (25%), Positives = 49/104 (47%)
Frame = -2
Query: 610 DKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDD 669
D++ +E+ N + + +DSD ++K + E QK+ + E+E S
Sbjct: 301 DEKSQEKPSEDTKTENQDTSVSEKRSDSDESQQKSDSDESQQKSDSD----ESEKKSDSA 134
Query: 670 RSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPKS 713
S+ +S SDE +KS+ + E ++ S ++ + ENK S
Sbjct: 133 ESEKKS-DSDESEKKSDSDETEKSSESNDNKQFDSDERENKSDS 5
Score = 30.4 bits (67), Expect = 5.0
Identities = 37/162 (22%), Positives = 67/162 (40%), Gaps = 2/162 (1%)
Frame = -2
Query: 130 QLKSQKSTKKDGRSKQVGNARKGTQK--TKTVHSEDGNSQQNVKNVSMESSPFKPHDAPP 187
++K Q + + S+Q + R + TK S + +VK E S K + P
Sbjct: 454 EVKEQATDPSNNNSQQFEDNRGDLSEDATKGDGSVTPDKNSDVKEKQEEKSDEKSQEKPS 275
Query: 188 STVNECDTSTKDKHMVTDYEVLQKSSTQEEPKPGSTTSVAYGVQERDQKYIKKWHLMYKH 247
DT T+++ D V +K S +E + S + + Q+ D +K +
Sbjct: 274 E-----DTKTENQ----DTSVSEKRSDSDESQQKSDSDESQ--QKSDSDESEKKSDSAES 128
Query: 248 AVLSNTGKCDNKVPLVEKEKEGGEEDNEGNNSYRNYSETDSD 289
S++ + + K E EK DN+ +S +++DSD
Sbjct: 127 EKKSDSDESEKKSDSDETEKSSESNDNKQFDSDERENKSDSD 2
>TC213762
Length = 465
Score = 32.0 bits (71), Expect = 1.7
Identities = 20/70 (28%), Positives = 32/70 (45%)
Frame = +3
Query: 1077 NSTIDRTGGIYSEGLNQKEQKMESGNGIVEERKEESVSKEVNKPNQKLSRNWSNLKKVVL 1136
+ ++ G I +E + Q EQ ++ + E KEE +S E KP + LKKV
Sbjct: 30 SEVVESLGSIINEIVPQDEQIPQAAVVAIGEEKEEKLSVENTKPEPSAKQGNYYLKKVFE 209
Query: 1137 LRRFIKALEK 1146
L+ + K
Sbjct: 210 LKDIVSLRNK 239
>TC210224 similar to UP|Q84T10 (Q84T10) Unknow protein, partial (19%)
Length = 7461
Score = 32.0 bits (71), Expect = 1.7
Identities = 35/154 (22%), Positives = 59/154 (37%), Gaps = 12/154 (7%)
Frame = +1
Query: 579 QKYIKKWHLMYKQAVLSNTGKYDNKLPVVGKDKEGREQGDAVFNGGNN---SSCHNYNET 635
+ ++K Y+ S + D + + + E F G + SS ++
Sbjct: 355 ESFLKNGKFRYRTIPSSQKKQLDERFNAITQRVESFSPVKKSFCGKHKRFMSSASEDEDS 534
Query: 636 DSDMDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEKSEGEREEMNAT 695
DS DE+ N+I+ Q P ++ S +R S Y S + G R +M
Sbjct: 535 DSSTDEQSNNIIKGSQSN------PSSQFTRSSERVSSCPYPSATEEKARLGVRSDMAGH 696
Query: 696 S---------FTETPKEAKKTENKPKSWSHLKKL 720
S F+E P++ +K EN + S KL
Sbjct: 697 SLVNSNLKKGFSEQPRKKRKFENATSTRSAPYKL 798
>TC229604 similar to UP|Q9M276 (Q9M276) Homeobox-leucine zipper protein
ATHB-12, partial (44%)
Length = 992
Score = 32.0 bits (71), Expect = 1.7
Identities = 24/118 (20%), Positives = 50/118 (42%), Gaps = 5/118 (4%)
Frame = +2
Query: 685 SEGEREEMNATSFTETPKEAKKTENKPKSWSHLKKLIMLKRFVKALDKVRNINPRRPREL 744
S G E + +S T + K+ N + +S + +K + + PR+ +L
Sbjct: 158 SAGVEAETHTSSSTTPSRSKKRNNNNTRRFSDEQ----IKSLETMFESETRLEPRKKLQL 325
Query: 745 PSDANFEGEKVFLNRQTSEERKKSEEWMLDYAL-----QKVISKLAPAQRQRVTLLIE 797
+ + +V + Q R KS++ DY + + S+ ++++ TLLI+
Sbjct: 326 ARELGLQPRQVAIWFQNKRARWKSKQLERDYGILQSNYNTLASRFEALKKEKQTLLIQ 499
>TC222232 similar to UP|Q6UEJ2 (Q6UEJ2) Mini-chromosome maintenance 7,
partial (18%)
Length = 445
Score = 32.0 bits (71), Expect = 1.7
Identities = 17/61 (27%), Positives = 30/61 (48%), Gaps = 4/61 (6%)
Frame = +1
Query: 631 NYNETDSD----MDEEKKNVIELVQKAFDEILLPETEDLSSDDRSKSRSYGSDELLEKSE 686
NY + D + + + + I + A DE++ TED + DD + SDE +E ++
Sbjct: 220 NYKDLDEEFLSRVTDNTRRYIGIFSNAIDELMPEPTEDFTDDDHDILMTQRSDEGVEGTD 399
Query: 687 G 687
G
Sbjct: 400 G 402
>TC216094 similar to UP|Q6SJQ9 (Q6SJQ9) TFIID component TAF2 (Fragment),
partial (20%)
Length = 2400
Score = 31.6 bits (70), Expect = 2.2
Identities = 33/146 (22%), Positives = 61/146 (41%)
Frame = +3
Query: 351 ESPKEVPKMESKQKSWSHLKKVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLN 410
E+PK+ P Q LK+ +LEK + + + P++ P +A E K
Sbjct: 1050 EAPKDQPCEAPTQVHLEALKEA--------SLEKPKEVFTEFPQEAPIEAPNEISKEADT 1205
Query: 411 RQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLVEAFETIRPVQDAENGPQTSATV 470
S ERK+ + KV A ++ +VE ++ + ++G +S +V
Sbjct: 1206 VSNSHERKRPIK-------IKVKQSSATSRADTDNQVVE--RSLGGRNEMDHGASSSVSV 1358
Query: 471 ESHANLIQSLDASSNHSKEEINDRRD 496
++ + SNH+ +E+N D
Sbjct: 1359 DAPQRNFAETVSISNHNIDEVNSWHD 1436
>TC206105 weakly similar to UP|Q7RH28 (Q7RH28) Mature-parasite-infected
erythrocyte surface antigen, partial (5%)
Length = 858
Score = 30.8 bits (68), Expect = 3.8
Identities = 26/107 (24%), Positives = 47/107 (43%)
Frame = +1
Query: 263 VEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHS 322
V +EK+ E E + + + ++ +EKK V +K E+++ E +++
Sbjct: 550 VTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKKEAEVIVEEKKEVEVTEEK 729
Query: 323 KSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWSHL 369
K E + +K IEE+ T TE KEV ++K S +
Sbjct: 730 K----EVEVTEGKKEVEVIEEKKETEVTEEKKEVEVEVREEKKESEV 858
>TC216791 similar to UP|O23144 (O23144) Proton pump interactor
(AT4g27500/F27G19_100), partial (25%)
Length = 1385
Score = 30.4 bits (67), Expect = 5.0
Identities = 26/108 (24%), Positives = 45/108 (41%), Gaps = 3/108 (2%)
Frame = +3
Query: 314 EDLSSEGHSKSRGNETDEVLLEKSGGK-IEERNTTTFTESPKEVPKMESK--QKSWSHLK 370
E L ++ SK R L K K + E + F KE P E + +K
Sbjct: 474 ETLPAQKESKKRSKNKGRDLKSKPESKDVAETDEYEFKNPQKEAPAKEPEIDPAKLKEMK 653
Query: 371 KVILLKRFVKALEKVRNINSRRPRQLPSDANFEAEKVLLNRQTSEERK 418
+ + + +ALE+ + + + + A EAEK L +R+ ++K
Sbjct: 654 REEEIAKAKQALERKKKLAEKAAAKAAIRAQKEAEKKLKDREKKAKKK 797
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 30.4 bits (67), Expect = 5.0
Identities = 27/103 (26%), Positives = 44/103 (42%)
Frame = +2
Query: 610 DKEGREQGDAVFNGGNNSSCHNYNETDSDMDEEKKNVIELVQKAFDEILLPETEDLSSDD 669
D++ E+GD V GG N ++ DSD DE++ E Q +++ TE L
Sbjct: 431 DEDDEEEGD-VQRGGEPDDDDNDSDDDSDDDEDEDEEDEEEQGEEEDL---GTEYLIRPL 598
Query: 670 RSKSRSYGSDELLEKSEGEREEMNATSFTETPKEAKKTENKPK 712
+ S + + GE EE + ++ +K E PK
Sbjct: 599 ETAEEEEASSDFEPEENGEEEEEDVDD-----EDCEKAEAPPK 712
>BU763591
Length = 389
Score = 30.4 bits (67), Expect = 5.0
Identities = 26/105 (24%), Positives = 46/105 (43%)
Frame = -1
Query: 263 VEKEKEGGEEDNEGNNSYRNYSETDSDMDDEKKNVIELVQKAFDEILLPEVEDLSSEGHS 322
V +EK+ E E + + + ++ +EKK V +K E+++ E +++
Sbjct: 308 VTEEKKEIEVTEEKKEAEVKEEKKEGEVTEEKKEVEVTEEKKEAEVIVEEKKEVEVTEEK 129
Query: 323 KSRGNETDEVLLEKSGGKIEERNTTTFTESPKEVPKMESKQKSWS 367
K E + +K IEE+ T TE KEV ++K S
Sbjct: 128 K----EVEVTEGKKEVEVIEEKKETEVTEEKKEVEVEVREEKKES 6
>TC204046 UP|Q39835 (Q39835) Extensin, complete
Length = 1627
Score = 30.4 bits (67), Expect = 5.0
Identities = 29/95 (30%), Positives = 44/95 (45%)
Frame = -1
Query: 931 SNLVVPEAPSNRLDEPTTDIKDVVEKDQLEKSEAPTSAVVESKNQLEKQGSTGLWFTVFK 990
S LVV E+ SNR +E + +VVE D + E VVES + +E++ S +
Sbjct: 601 SALVVVESGSNREEEVSA*--EVVESDSSMEEEVSA*EVVESDSSMEEEVSVSV------ 446
Query: 991 HMVSDMTENNSKTSTDVADEKDSKYEDITTREISV 1025
+V + + S V DS E+ E+SV
Sbjct: 445 -VVGSDSNMEEEVSVSVVVGSDSSMEE----EVSV 356
>TC217749 weakly similar to UP|Q6Z5M6 (Q6Z5M6) Transducin / WD-40 repeat
protein-like, partial (22%)
Length = 1079
Score = 30.0 bits (66), Expect = 6.5
Identities = 33/124 (26%), Positives = 52/124 (41%), Gaps = 1/124 (0%)
Frame = -1
Query: 748 ANFEGEKVFLNRQTSEERKKSEEWMLDYA-LQKVISKLAPAQRQRVTLLIEAFETLRPIQ 806
ANF G V +++ R +S W+ + K+ +K Q ET+
Sbjct: 758 ANFLGVSVLMSQDNCCRRDRSTPWLNGSGTIAKIRAKST*EHNQ---------ETIETSL 606
Query: 807 DAENGLRSSATVESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDKASDNPMPELC 866
+ +S+ + SL N SL SSV+S K LL + + +T+E AS P
Sbjct: 605 L*RAPVNASSNLPSLINLDSSLVGSSVISCKPLLICATKGSAATLELVAIASSFRWPSAN 426
Query: 867 KPIK 870
P+K
Sbjct: 425 CPLK 414
>TC204685 similar to UP|RK34_SPIOL (P82244) 50S ribosomal protein L34,
chloroplast precursor, partial (57%)
Length = 783
Score = 30.0 bits (66), Expect = 6.5
Identities = 24/99 (24%), Positives = 45/99 (45%)
Frame = +3
Query: 758 NRQTSEERKKSEEWMLDYALQKVISKLAPAQRQRVTLLIEAFETLRPIQDAENGLRSSAT 817
NR+ E++KK+++W++ +L I+ AP + L +P + N R+ +T
Sbjct: 24 NREVKEKKKKNQKWIM-ASLSLCIAMTAPTPSASLILTAP-----KPSSLSLNLTRTCST 185
Query: 818 VESLENPLQSLDASSVLSAKTLLGKVSFSNDSTMEFSDK 856
S L + S+LS + + +S D T F +
Sbjct: 186 ARSPLLHCSFLSSPSLLSPPSSISGLSLGLDLTSNFGTR 302
>TC217093
Length = 1829
Score = 30.0 bits (66), Expect = 6.5
Identities = 36/175 (20%), Positives = 79/175 (44%), Gaps = 17/175 (9%)
Frame = +2
Query: 488 KEEINDRRDFEVTERARNDKNMDA-CKKNDESATVKSTATKAV-KFPVCDTGIMEEEVTA 545
+E+I ++ + ++ ++ +N +A K+ ++ T K+T + K P + + +T
Sbjct: 1040 EEKIQEKEAEKTNQQEKSKENQEAEIKQLRKTMTFKATPMPSFYKEPPPKVELKKIPITR 1219
Query: 546 EGEYKV-QEKSIVKEDLKHGTSTTDVPYGVQERDQKYIKKW-----HLMYKQAVLSNTGK 599
K+ + K + + +S++ P+G Q+++ K ++ K+ + + K
Sbjct: 1220 PKSPKLGRHKGSAVNNSEDKSSSS--PHGKQQQNDSTKAKAKGSNKEVIPKKPIRKSQAK 1393
Query: 600 YDNKLPVVGKD-----KEGREQGDAVFNGGNNSSCH----NYNETDSDMDEEKKN 645
++ + K K + DA GNN CH N +E +DM++E KN
Sbjct: 1394 LQSQESAIRKTEKDSVKPTKVDQDAKAGTGNNEECHDPSVNNSEYQNDMEQESKN 1558
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,697,656
Number of Sequences: 63676
Number of extensions: 525916
Number of successful extensions: 2860
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 2681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2848
length of query: 1222
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1114
effective length of database: 5,762,624
effective search space: 6419563136
effective search space used: 6419563136
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC125478.8


