

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.4 + phase: 0
(260 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
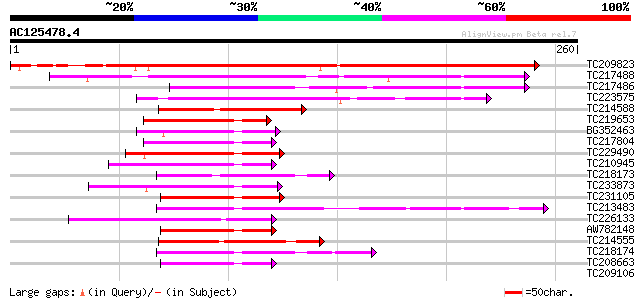
Score E
Sequences producing significant alignments: (bits) Value
TC209823 279 8e-76
TC217488 74 7e-14
TC217486 65 4e-11
TC223575 55 2e-08
TC214588 homologue to UP|Q71SQ1 (Q71SQ1) MYC1, partial (35%) 46 2e-05
TC219653 similar to GB|AAL79583.1|18958020|AY079033 AT3g25710/K1... 45 4e-05
BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thalian... 44 7e-05
TC217804 44 7e-05
TC229490 similar to UP|Q8S9M5 (Q8S9M5) At2g41130/T3K9.10, partia... 44 1e-04
TC210945 similar to GB|BAC56979.1|28372351|AB103113 PIF3 like ba... 44 1e-04
TC218173 similar to UP|Q41102 (Q41102) Phaseolin G-box binding p... 44 1e-04
TC233873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein famil... 43 1e-04
TC231105 43 1e-04
TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, parti... 43 1e-04
TC226133 similar to UP|Q6ZBQ2 (Q6ZBQ2) BHLH protein family-like,... 43 2e-04
AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thali... 42 2e-04
TC214555 similar to GB|AAO23602.1|27764962|BT003037 AT3g57800/T1... 42 4e-04
TC218174 similar to UP|Q41102 (Q41102) Phaseolin G-box binding p... 41 6e-04
TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacti... 40 8e-04
TC209106 similar to UP|Q41101 (Q41101) Phaseolin G-box binding p... 40 0.001
>TC209823
Length = 1092
Score = 279 bits (714), Expect = 8e-76
Identities = 163/251 (64%), Positives = 189/251 (74%), Gaps = 8/251 (3%)
Frame = +1
Query: 1 MVA-FCPPQFSYSNMGWLLEELEPESLISHKEKNYASLEYSLPYHQFSSPKEHVEIE--- 56
MVA F PP FS GW LEE EP S S +YS PY QF +P+ +E+E
Sbjct: 49 MVALFSPPVFS--TKGWFLEE-EPLSYD-------VSSDYSFPY-QFFAPQTQIELEIER 195
Query: 57 -RPPSPK--LMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKY 113
PSP+ M KKL+HNASERDRRKK+N L+SSLRSLLPG DQTKKMSIP T+SRVLKY
Sbjct: 196 SSAPSPEDPAMVKKLSHNASERDRRKKVNDLVSSLRSLLPGPDQTKKMSIPATVSRVLKY 375
Query: 114 IPDLQKQVQGLTKKKEELLSRISHRQEY-AVNKESQRKKIPNYNSAFVVSTSRLNDTELV 172
IP+LQ QVQ LTKKKEELL RIS + +VNKESQR+ I ++NS F VSTSRLND E V
Sbjct: 376 IPELQHQVQALTKKKEELLCRISKNLKGDSVNKESQRR-ISHHNSDFAVSTSRLNDCEAV 552
Query: 173 IHISSYEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLI 232
+HISSYEA+K PLS+IL CLENNGL LLN+SSS+TFGGR+FYNLHFQV+KT R E + L
Sbjct: 553 VHISSYEAHKAPLSDILQCLENNGLYLLNASSSETFGGRVFYNLHFQVEKTHRLESEILT 732
Query: 233 QKLSSIYEKQQ 243
+KL SIYEKQ+
Sbjct: 733 EKLLSIYEKQR 765
>TC217488
Length = 977
Score = 73.9 bits (180), Expect = 7e-14
Identities = 61/222 (27%), Positives = 109/222 (48%), Gaps = 2/222 (0%)
Frame = +2
Query: 19 EELEPESLISHKEKNY-ASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKKLNHNASERDR 77
++++ L+ + +K S L YH+ + + E KK+ H E+ R
Sbjct: 203 QDIDQSPLVGYSDKKLRTSRPKKLFYHEADTCHANSNQE-------YTKKMVHREIEKQR 361
Query: 78 RKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLSRISH 137
R+++ +L +SLRSLLP + K SI ++ + YI LQK ++ L+ K+++L + S
Sbjct: 362 RQEMATLHASLRSLLPLDFIKGKRSISDQMNEAVNYINHLQKNIKELSDKRDKLKKKPS- 538
Query: 138 RQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELV-IHISSYEANKIPLSEILMCLENNG 196
+N + + NY A T N V I IS + ++PLS++L + G
Sbjct: 539 ----IINSSPEDHE--NYKHASSGFTVHQNSGGAVGIEISGFSEEEVPLSKLLELVFEEG 700
Query: 197 LLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSI 238
L ++N S+K GRL ++L +VD + + +L +K S +
Sbjct: 701 LEVVNCLSTKV-NGRLLHSLQCEVDNSGSVDLSELRRKFSDV 823
>TC217486
Length = 753
Score = 64.7 bits (156), Expect = 4e-11
Identities = 46/166 (27%), Positives = 91/166 (54%), Gaps = 1/166 (0%)
Frame = +2
Query: 74 ERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKKEELLS 133
ER RR+++ +L +SLRSLLP K SI +S + YI LQK ++ L+ K+++L
Sbjct: 8 ERQRRQEMATLHASLRSLLPLRFIKGKRSISDQMSEAVNYINHLQKNIKELSDKRDKL-- 181
Query: 134 RISHRQEYAVNKESQ-RKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPLSEILMCL 192
+++ ++N + + + +S F + + + + I IS + ++PLS++L +
Sbjct: 182 ----KKKPSINSTPEGHENCKHVSSGFTIHQN--SGGAVGIEISGFSEEEVPLSKLLKLV 343
Query: 193 ENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSI 238
L +++ S+K GRL ++L +VD + + +L +K S++
Sbjct: 344 LEERLEVVSCLSTKV-NGRLLHSLQCEVDNSDSVDLSELRRKFSNV 478
>TC223575
Length = 802
Score = 55.5 bits (132), Expect = 2e-08
Identities = 46/164 (28%), Positives = 80/164 (48%), Gaps = 1/164 (0%)
Frame = +3
Query: 59 PSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQ 118
PSP + +K+ E++RR ++ +L S L SLLP + + MS+P I + YI L+
Sbjct: 45 PSPTKVERKI----VEKNRRSQMKNLYSELNSLLPTRNPKEAMSLPDQIDEAINYIKSLE 212
Query: 119 KQVQGLTKKKEELLSRISHRQEYAVNKESQRK-KIPNYNSAFVVSTSRLNDTELVIHISS 177
+V+ +KKE L R R + + E+Q K PN + T L + L + S
Sbjct: 213 TKVKLEQEKKERLKERKRTRGGCSSSSEAQGSLKSPNIQ---IHETGNLLEVILTCGVDS 383
Query: 178 YEANKIPLSEILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVD 221
+ EI+ L + ++N++SS G + + +H +V+
Sbjct: 384 ----QFMFCEIIRILHEENVEVINANSSMV-GDLVIHVVHGEVE 500
>TC214588 homologue to UP|Q71SQ1 (Q71SQ1) MYC1, partial (35%)
Length = 837
Score = 45.8 bits (107), Expect = 2e-05
Identities = 24/68 (35%), Positives = 41/68 (60%)
Frame = +3
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
NH+ +ER RR+KI+ + L+ L+PG K + + ++ Y+ LQ+QV+ L+ K
Sbjct: 417 NHSLAERARREKISERMKILQDLVPG--CNKVIDKAYVLDEIINYVQSLQRQVEFLSMKL 590
Query: 129 EELLSRIS 136
E + SR+S
Sbjct: 591 EAVSSRLS 614
>TC219653 similar to GB|AAL79583.1|18958020|AY079033 AT3g25710/K13N2_1
{Arabidopsis thaliana;} , partial (47%)
Length = 1346
Score = 44.7 bits (104), Expect = 4e-05
Identities = 21/59 (35%), Positives = 41/59 (68%)
Frame = +2
Query: 62 KLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQ 120
K +A +H+ +ER RR++IN+ ++ LRSLLP +T K S+ ++ V++++ +L++Q
Sbjct: 515 KALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASL---LAEVIQHVKELKRQ 682
>BG352463 similar to GP|11245494|gb| SPATULA {Arabidopsis thaliana}, partial
(19%)
Length = 471
Score = 43.9 bits (102), Expect = 7e-05
Identities = 24/70 (34%), Positives = 41/70 (58%), Gaps = 4/70 (5%)
Frame = +3
Query: 59 PSPKLMAKKLN----HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYI 114
P P+ +K+ HN SE+ RR +IN + +L++L+P ++T K S+ + ++Y+
Sbjct: 18 PPPRSSSKRSRAAEFHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASM---LDEAIEYL 188
Query: 115 PDLQKQVQGL 124
LQ QVQ L
Sbjct: 189 KQLQLQVQYL 218
>TC217804
Length = 1578
Score = 43.9 bits (102), Expect = 7e-05
Identities = 23/61 (37%), Positives = 35/61 (56%)
Frame = +1
Query: 62 KLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQV 121
++ A + H SER RR KIN + L+ L+P +T K S+ + V++Y+ LQ QV
Sbjct: 799 RIKANSVVHKQSERRRRDKINQRMKELQKLVPNSSKTDKASM---LDEVIQYMKQLQAQV 969
Query: 122 Q 122
Q
Sbjct: 970 Q 972
>TC229490 similar to UP|Q8S9M5 (Q8S9M5) At2g41130/T3K9.10, partial (56%)
Length = 1006
Score = 43.5 bits (101), Expect = 1e-04
Identities = 24/75 (32%), Positives = 46/75 (61%), Gaps = 2/75 (2%)
Frame = +1
Query: 54 EIERPPS--PKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVL 111
+I PS + +A NH +E+ RR++INS + LR+LLP +T K S+ +++V+
Sbjct: 292 QITETPSHHDRALAAMKNHKEAEKRRRERINSHLDHLRTLLPCNSKTDKASL---LAKVV 462
Query: 112 KYIPDLQKQVQGLTK 126
+ + +L++Q +T+
Sbjct: 463 QRVKELKQQTSEITE 507
>TC210945 similar to GB|BAC56979.1|28372351|AB103113 PIF3 like basic Helix
Loop Helix protein {Arabidopsis thaliana;} , partial
(19%)
Length = 432
Score = 43.5 bits (101), Expect = 1e-04
Identities = 25/77 (32%), Positives = 41/77 (52%)
Frame = +2
Query: 46 FSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPV 105
F SP+ ++ S K HN SER RR +IN + +L+ L+P +++ K S+
Sbjct: 41 FESPEAKKQVHGSTSTKRSHAAEVHNLSERSRRDRINEKMKALQELIPRCNKSDKASM-- 214
Query: 106 TISRVLKYIPDLQKQVQ 122
+ ++Y+ LQ QVQ
Sbjct: 215 -LDEAIEYLKSLQLQVQ 262
>TC218173 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (28%)
Length = 867
Score = 43.5 bits (101), Expect = 1e-04
Identities = 26/82 (31%), Positives = 43/82 (51%)
Frame = +3
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P KM + + YI +L+ ++Q L
Sbjct: 27 LNHVEAERQRREKLNQRFYALRAVVP---NVSKMDXASLLGDAISYITELKSKLQTLESD 197
Query: 128 KEELLSRISHRQEYAVNKESQR 149
K+ L H+Q V KE ++
Sbjct: 198 KDVL-----HKQLEGVKKELEK 248
>TC233873 weakly similar to UP|Q6ZJC8 (Q6ZJC8) BHLH protein family-like,
partial (25%)
Length = 817
Score = 43.1 bits (100), Expect = 1e-04
Identities = 29/92 (31%), Positives = 52/92 (56%), Gaps = 3/92 (3%)
Frame = +3
Query: 37 LEYSLPYHQFSSPKEHVEIERPPSP---KLMAKKLNHNASERDRRKKINSLISSLRSLLP 93
L++ + Q S K+ VE++ S K +H +ER RR++INS +S+LR+LLP
Sbjct: 39 LDHDAGHAQSSLIKKAVELDGARSKTERKSTEACKSHREAERRRRQRINSHLSTLRTLLP 218
Query: 94 GEDQTKKMSIPVTISRVLKYIPDLQKQVQGLT 125
++ K S+ + V++++ L+KQ +T
Sbjct: 219 NAAKSDKASL---LGEVVEHVKRLRKQADDVT 305
>TC231105
Length = 449
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/57 (36%), Positives = 35/57 (60%)
Frame = +2
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTK 126
HN SER RR KIN + +L+ L+P ++ K S+ + V++Y+ LQ Q+Q + +
Sbjct: 185 HNQSERKRRDKINQRMKTLQKLVPNSSKSDKASM---LDEVIEYLKQLQAQLQMINR 346
>TC213483 similar to UP|Q8LPT0 (Q8LPT0) At2g46510/F13A10.4, partial (17%)
Length = 914
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/180 (21%), Positives = 80/180 (44%)
Frame = +2
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LRS++P + K S+ + + YI +LQ +V+ + +
Sbjct: 152 LNHVEAERQRREKLNQRFYALRSVVPNISKMDKASL---LGDTIAYINELQAKVKIMEAE 322
Query: 128 KEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLNDTELVIHISSYEANKIPLSE 187
+E S + +E + V + D E+++ +S N PLS+
Sbjct: 323 RERFESISNQEKEAPAD----------------VDIQAVQDDEVIVRVSCPLDNH-PLSK 451
Query: 188 ILMCLENNGLLLLNSSSSKTFGGRLFYNLHFQVDKTQRYECDDLIQKLSSIYEKQQNNHL 247
++ + ++ S + +F+ + +++ D KL +++ ++N L
Sbjct: 452 VIQTFYQTQISVVESKLASA-NDAIFHTFVIKSQGSEQLTKD----KLIAVFSPTESNSL 616
>TC226133 similar to UP|Q6ZBQ2 (Q6ZBQ2) BHLH protein family-like, partial
(23%)
Length = 1519
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/95 (29%), Positives = 44/95 (45%)
Frame = +1
Query: 28 SHKEKNYASLEYSLPYHQFSSPKEHVEIERPPSPKLMAKKLNHNASERDRRKKINSLISS 87
S +E Y E+ Q +S + + K A + H+ +E+ RR KIN
Sbjct: 277 SQEEDEYEEEEFGSSKKQGTSSAPNANKDGKAIDKASAIRSKHSVTEQRRRSKINERFQI 456
Query: 88 LRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQ 122
LR L+P DQ K + V++Y+ LQ++VQ
Sbjct: 457 LRDLIPHSDQ--KRDTASFLLEVIEYVQYLQEKVQ 555
>AW782148 homologue to GP|11245494|gb| SPATULA {Arabidopsis thaliana},
partial (17%)
Length = 381
Score = 42.4 bits (98), Expect = 2e-04
Identities = 20/53 (37%), Positives = 34/53 (63%)
Frame = +1
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQ 122
HN SE+ RR +IN + +L++L+P ++T K S+ + ++Y+ LQ QVQ
Sbjct: 229 HNLSEKRRRSRINEKMKALQNLIPNSNKTDKASM---LDEAIEYLKQLQLQVQ 378
>TC214555 similar to GB|AAO23602.1|27764962|BT003037 AT3g57800/T10K17_10
{Arabidopsis thaliana;} , partial (22%)
Length = 737
Score = 41.6 bits (96), Expect = 4e-04
Identities = 22/76 (28%), Positives = 47/76 (60%)
Frame = +2
Query: 69 NHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKKK 128
+H+ +ER RR+KIN+ + L+ L+PG D K + + ++ ++ LQ+QV+ L+ K
Sbjct: 2 SHSLAERARREKINARMKLLQELVPGCD--KISGTAMVLDEIINHVQSLQRQVEILSMK- 172
Query: 129 EELLSRISHRQEYAVN 144
L+ ++ R +++++
Sbjct: 173 ---LAAVNPRMDFSLD 211
>TC218174 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (49%)
Length = 1345
Score = 40.8 bits (94), Expect = 6e-04
Identities = 29/101 (28%), Positives = 52/101 (50%)
Frame = +3
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P + K S+ + + YI +L+ ++Q L
Sbjct: 429 LNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASL---LGDAISYITELKSKLQTLESD 599
Query: 128 KEELLSRISHRQEYAVNKESQRKKIPNYNSAFVVSTSRLND 168
K+ + +Q V KE + K N +S ++S N+
Sbjct: 600 KDGM-----QKQLEGVKKELE-KTTENGSSNHAGNSSSCNN 704
>TC208663 similar to UP|PIF4_ARATH (Q8W2F3) Phytochrome-interacting factor 4
(Basic helix-loop-helix protein 9) (bHLH9) (AtbHLH009)
(Short under red-light 2), partial (19%)
Length = 1102
Score = 40.4 bits (93), Expect = 8e-04
Identities = 20/53 (37%), Positives = 32/53 (59%)
Frame = +1
Query: 70 HNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQ 122
HN SER RR +IN + +L+ L+P +T K S+ + ++Y+ LQ Q+Q
Sbjct: 274 HNLSERRRRDRINEKMKALQQLIPHSSKTDKASM---LEEAIEYLKSLQLQLQ 423
>TC209106 similar to UP|Q41101 (Q41101) Phaseolin G-box binding protein PG1,
partial (40%)
Length = 1101
Score = 40.0 bits (92), Expect = 0.001
Identities = 32/123 (26%), Positives = 62/123 (50%), Gaps = 10/123 (8%)
Frame = +3
Query: 68 LNHNASERDRRKKINSLISSLRSLLPGEDQTKKMSIPVTISRVLKYIPDLQKQVQGLTKK 127
LNH +ER RR+K+N +LR+++P + K S+ + + YI +L+ ++ L +
Sbjct: 306 LNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASL---LGDAILYINELKSKLNVLDSE 476
Query: 128 KEELLSRI-SHRQEYAVNKESQRK---------KIPNYNSAFVVSTSRLNDTELVIHISS 177
K EL ++ S ++E + ++ P+ + +TS+L D EL + I
Sbjct: 477 KTELEKQLDSTKKELELATKNPPPPPPPPPPPGPPPSNSVEPKKTTSKLADLELEVKIIG 656
Query: 178 YEA 180
++A
Sbjct: 657 WDA 665
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,747,733
Number of Sequences: 63676
Number of extensions: 163264
Number of successful extensions: 717
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 712
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 715
length of query: 260
length of database: 12,639,632
effective HSP length: 95
effective length of query: 165
effective length of database: 6,590,412
effective search space: 1087417980
effective search space used: 1087417980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC125478.4


