

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125477.15 - phase: 0
(738 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
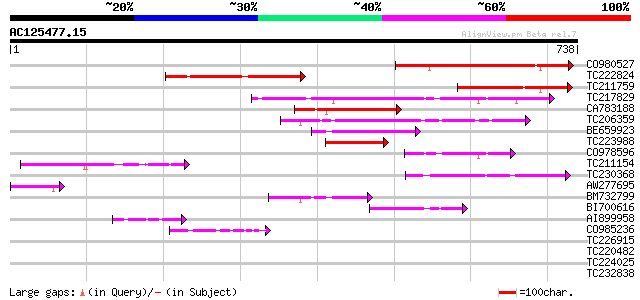
Score E
Sequences producing significant alignments: (bits) Value
CO980527 346 2e-95
TC222824 weakly similar to GB|AAA98791.1|212879|CHKVITB vitellog... 231 9e-61
TC211759 213 2e-55
TC217829 similar to UP|Q945L7 (Q945L7) AT4g35240/F23E12_200, par... 185 7e-47
CA783188 118 1e-26
TC206359 similar to UP|Q9LY33 (Q9LY33) BZIP protein, partial (84%) 115 7e-26
BE659923 99 9e-21
TC223988 similar to GB|AAS99727.1|46518491|BT012583 At2g27090 {A... 95 1e-19
CO978596 92 6e-19
TC211154 similar to GB|AAS99727.1|46518491|BT012583 At2g27090 {A... 79 9e-15
TC230368 69 1e-11
AW277695 65 8e-11
BM732799 58 1e-08
BI700616 54 2e-07
AI899958 48 1e-05
CO985236 44 2e-04
TC226915 similar to UP|Q8LDZ7 (Q8LDZ7) Cinnamoyl CoA reductase-l... 38 0.019
TC220482 similar to GB|AAS99727.1|46518491|BT012583 At2g27090 {A... 37 0.024
TC224025 weakly similar to UP|Q6HXC4 (Q6HXC4) Conserved domain p... 37 0.024
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 35 0.16
>CO980527
Length = 781
Score = 346 bits (887), Expect = 2e-95
Identities = 178/243 (73%), Positives = 204/243 (83%), Gaps = 12/243 (4%)
Frame = -3
Query: 503 EELHSQLLELLQGLAKMWKVMAECHQTQKQTLDEAKILIAGID----ARKQSSMSITDPY 558
+ELH Q LEL+QGL +MWKVMAECHQTQK+TLDEAKIL+AG ARKQSS+S+TDP
Sbjct: 779 KELHPQXLELVQGLERMWKVMAECHQTQKRTLDEAKILLAGTPSKSRARKQSSISMTDPN 600
Query: 559 RLARSASNLETELRNWRNTFESWITSQKSYIHALTGWLLRCMRCEPDASKLICSPRRSSS 618
RLARSASNLE ELRNWRN FESWITSQ+SYIHALTGWLLRCMR EPD SKL CSPRRSSS
Sbjct: 599 RLARSASNLEFELRNWRNAFESWITSQRSYIHALTGWLLRCMRFEPDVSKLPCSPRRSSS 420
Query: 619 THPLFGLCIQWSRRLDALEETAVLDSIDFFAAELGSFYAQQLREDSAQNA--TVGSNVNM 676
THPLFGLC+QWSRRLDA++E AVLD +DFFAA +GS YA QLREDS +N+ + SN NM
Sbjct: 419 THPLFGLCVQWSRRLDAIQEKAVLDGLDFFAAGMGSLYAHQLREDSRRNSFGSKQSNGNM 240
Query: 677 EMVEVAKVGEEVM------EVAVKVLCGGMSGAMSSMSEFAIDSAKGYNELVKQWENAKL 730
EMVEV +V EEVM EVA+KVLC GMS A+SS++EFA+DS++GYNE+VKQW+N K
Sbjct: 239 EMVEVGEV-EEVMAPEKLAEVAIKVLCAGMSVAISSLAEFALDSSEGYNEVVKQWDNGKC 63
Query: 731 QQT 733
Q T
Sbjct: 62 QNT 54
>TC222824 weakly similar to GB|AAA98791.1|212879|CHKVITB vitellogenin {Gallus
gallus;} , partial (17%)
Length = 703
Score = 231 bits (589), Expect = 9e-61
Identities = 122/182 (67%), Positives = 144/182 (79%)
Frame = +3
Query: 204 DDENRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDK 263
DDE RGLRKVREEEGIPDLE++E + E+ +KRNVAEER KIDVN S+EEV VEDV +
Sbjct: 6 DDEIRGLRKVREEEGIPDLEEDETEPEE--FAIKRNVAEERAKIDVNPSQEEVAVEDVYE 179
Query: 264 HKEEEKEKGTDAETETAQEISDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAKEKT 323
H+EEE+E+ T AET A E+SDS+ NG ECFQ SK+Q+ G +EMA +EAKE+T
Sbjct: 180 HEEEEEEEATGAETGIANEVSDSQANGSECFQASKAQTVG------QEMATGNQEAKEET 341
Query: 324 PGFTVYVNRRPENMAEVIKDLESQFTVVCNAANDVSALLEAKKAQYLSPSNELSASKLLN 383
PGFTVYVNRRP +M EVI+DLE+QFT++CNAANDVSALLEAKKAQYLS SNELS L
Sbjct: 342 PGFTVYVNRRPTSMVEVIQDLEAQFTIICNAANDVSALLEAKKAQYLSTSNELSGLLKLE 521
Query: 384 PV 385
V
Sbjct: 522 SV 527
>TC211759
Length = 798
Score = 213 bits (543), Expect = 2e-55
Identities = 109/158 (68%), Positives = 127/158 (79%), Gaps = 8/158 (5%)
Frame = +2
Query: 583 TSQKSYIHALTGWLLRCMRCEPDASKLICSPRRSSSTHPLFGLCIQWSRRLDALEETAVL 642
T Q+SYIHALTGWLLRC+RCE D SKL CSPRRSS THPLFGLC+QWSRRLDAL+ETAVL
Sbjct: 11 TRQRSYIHALTGWLLRCVRCEHDPSKLACSPRRSSGTHPLFGLCVQWSRRLDALQETAVL 190
Query: 643 DSIDFFAAELGSFYAQQLREDSAQNATVGSNVNMEMVEVAKVG--EEVM------EVAVK 694
D IDFFAA +GS YAQQLRE++ +N GS + E++E+ +VG EEVM EVA+K
Sbjct: 191 DGIDFFAAGIGSLYAQQLREETRRNPD-GSKEHGEIMEMLEVGQVEEVMNTEKLAEVAIK 367
Query: 695 VLCGGMSGAMSSMSEFAIDSAKGYNELVKQWENAKLQQ 732
VLC GMS AM SM+EFA+D A+GYNEL K+WEN LQQ
Sbjct: 368 VLCAGMSTAMRSMAEFAVDYAEGYNELAKRWENVNLQQ 481
>TC217829 similar to UP|Q945L7 (Q945L7) AT4g35240/F23E12_200, partial (68%)
Length = 1872
Score = 185 bits (469), Expect = 7e-47
Identities = 132/411 (32%), Positives = 208/411 (50%), Gaps = 16/411 (3%)
Frame = +1
Query: 315 DTEEAKEKTPGFTVYVNRRP--ENMAEVIKDLESQFTVVCNAANDVSALLEAKKAQYLSP 372
D +AKE + RRP N EV K+++ F ++ ++ +LE K +
Sbjct: 58 DGNKAKEHA----AFRTRRPGSRNPLEVAKEIQILFQRASDSGAQIAKILEVGKLPHNRK 225
Query: 373 SNELSAS-KLLNPVALFRSSPSKIITNFSKTRDEVYDDPSEEQCVFSV-------SHQST 424
AS K+L VA PS + + + + + S F+V + ST
Sbjct: 226 HAAYQASSKMLQVVA-----PSLSLVSSQPSTSKDAESASAANMDFNVDLTTGARNLSST 390
Query: 425 LDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDIKGEEPSSVDKTRAAIRDLHTQIT 484
L +L WEKKL+ EVK+ ++R+ +++KC +L+ D +G + VD TR IR+L T+I
Sbjct: 391 LQKLLLWEKKLFNEVKAEEKMRVMHDRKCHKLKRLDDRGADFHKVDSTRTLIRNLSTKIR 570
Query: 485 VSIHSVEAISRRIETLRDEELHSQLLELLQGLAKMWKVMAECHQTQKQTLDEAKILIAGI 544
++I V+ IS I +RDEEL QL EL+QGL +MWK M ECH Q + + EA+IL I
Sbjct: 571 MAIQVVDKISMTINKIRDEELWPQLKELIQGLTRMWKSMLECHHDQCEAIREARIL-GSI 747
Query: 545 DARKQSSMSITDPYRLARSASNLETELRNWRNTFESWITSQKSYIHALTGWLLRCMRCEP 604
+RK+S S ++ LE EL NW F WI++QK Y+ AL WLL+C+ EP
Sbjct: 748 GSRKKSGDS------HLQATKQLEQELINWTFQFSGWISAQKGYVRALNNWLLKCLLYEP 909
Query: 605 DASK---LICSPRRSSSTHPLFGLCIQWSRRLDALEETAVLDSIDFFAAELGSFYAQ--- 658
+ + + SP R + +F +C QWS+ LD + E V+DS+ F + + Q
Sbjct: 910 EETPDGIVPFSPGRIGAPQ-IFVICNQWSQALDRISEKEVVDSMHVFTMSVLQIWEQDKL 1086
Query: 659 QLREDSAQNATVGSNVNMEMVEVAKVGEEVMEVAVKVLCGGMSGAMSSMSE 709
++ +N + V + K+ +++ + KV+ G S+SE
Sbjct: 1087EMHRQVMKNKDLERKVRNMDRDDQKLQKQIQALERKVVLVSGEGKGLSVSE 1239
>CA783188
Length = 454
Score = 118 bits (295), Expect = 1e-26
Identities = 66/144 (45%), Positives = 98/144 (67%), Gaps = 5/144 (3%)
Frame = +1
Query: 371 SPSNELSASKLLN--PVALFRSSPSKIITNFSKTRDEVYDDPS---EEQCVFSVSHQSTL 425
SP + A K+++ A RSS S+ +KT++++ D S EE C+ + SH STL
Sbjct: 25 SPVFQEPAQKIISWKRTASSRSSSSRNALA-TKTKEDIDDSGSDFVEEFCMIAGSHSSTL 201
Query: 426 DRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDIKGEEPSSVDKTRAAIRDLHTQITV 485
DRLYAWE+KLY EVK+ +R Y++KC QLR+ K + +DKTR+ ++DLH+++TV
Sbjct: 202 DRLYAWERKLYDEVKASEFIRKDYDRKCHQLRHQFAKDQGTHVIDKTRSVVKDLHSRLTV 381
Query: 486 SIHSVEAISRRIETLRDEELHSQL 509
+I+SV++IS+RIE +RDEEL QL
Sbjct: 382 AIYSVDSISKRIERMRDEELLPQL 453
>TC206359 similar to UP|Q9LY33 (Q9LY33) BZIP protein, partial (84%)
Length = 1541
Score = 115 bits (288), Expect = 7e-26
Identities = 92/332 (27%), Positives = 160/332 (47%), Gaps = 7/332 (2%)
Frame = +2
Query: 353 NAANDVSALLEAKKAQYLSPSNEL------SASKLLNPVALFRSSPSKIITNFSKTRDEV 406
+A + VS +LE +AQ +L S+S L N + + S P + T
Sbjct: 14 SAGDQVSEMLEISRAQLDRSFKQLRKTVYHSSSILSNLSSSWTSKPPLAVKYRLDTGS-- 187
Query: 407 YDDPSEEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDIKGEEP 466
D+P + + S TL+RL AWEKKLY+EVK+ V++ +EKK L+ + KGE+
Sbjct: 188 LDEPGGPKSLCS-----TLERLLAWEKKLYEEVKAREGVKIEHEKKLSALQTQEYKGEDE 352
Query: 467 SSVDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQLLELLQGLAKMWKVMAEC 526
+ + KT+A+I L + I V+ +V S LRD +L QL++L+ G MW+ M
Sbjct: 353 AKIFKTKASINRLQSLIAVTSQAVSTTSTATIGLRDSDLVPQLVDLIHGFMYMWRSMHHY 532
Query: 527 HQTQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNWRNTFESWITSQK 586
H+ Q + + + G+ R S ++ +R ++ +LE+ + W ++F I Q+
Sbjct: 533 HEIQSNIVQQ----VRGLVNRSSRGDSTSELHR--QATRDLESAVSAWHSSFCRLIKFQR 694
Query: 587 SYIHALTGWL-LRCMRCEPDASKLICSPRRSSSTHPLFGLCIQWSRRLDALEETAVLDSI 645
+I +L GW L + D + R +S T+ F +W LD + +T ++I
Sbjct: 695 DFILSLHGWFKLSLVPVHNDN----INGRETSETYQFFD---EWKLALDRVPDTVASEAI 853
Query: 646 DFFAAELGSFYAQQLREDSAQNATVGSNVNME 677
F + ++Q+ E + T ++ +E
Sbjct: 854 KSFINVVHVISSKQVEELKIKKRTETASKELE 949
>BE659923
Length = 684
Score = 98.6 bits (244), Expect = 9e-21
Identities = 50/141 (35%), Positives = 80/141 (56%)
Frame = +2
Query: 394 KIITNFSKTRDEVYDDPSEEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEKKC 453
K I N +D+ D E +H + LD+L AWEKKLY EVK+G ++ Y++K
Sbjct: 29 KGIPNLDDGKDDFDSDEHE-------THATILDKLLAWEKKLYDEVKAGELMKFEYQRKV 187
Query: 454 LQLRNHDIKGEEPSSVDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQLLELL 513
L +G +++K +A + LHT+ V + S+++ I LRDE+L+ +L++L+
Sbjct: 188 AALNKLKKRGTNSEALEKAKAVVSHLHTRYIVDMQSLDSTVSEINRLRDEQLYPRLVQLV 367
Query: 514 QGLAKMWKVMAECHQTQKQTL 534
G+A MWK M E H Q +T+
Sbjct: 368 DGMATMWKTMLEHHVKQSETV 430
>TC223988 similar to GB|AAS99727.1|46518491|BT012583 At2g27090 {Arabidopsis
thaliana;} , partial (16%)
Length = 415
Score = 94.7 bits (234), Expect = 1e-19
Identities = 44/81 (54%), Positives = 59/81 (72%)
Frame = +1
Query: 412 EEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEKKCLQLRNHDIKGEEPSSVDK 471
+ C+ S SH STLDRLYAWE+KLY EVK+ VR Y+ KC LR + KGE+ S+VDK
Sbjct: 172 DNSCMISGSHASTLDRLYAWERKLYDEVKASEIVRKEYDMKCKFLRQLESKGEKTSTVDK 351
Query: 472 TRAAIRDLHTQITVSIHSVEA 492
TRA ++DLH++I V+IH + +
Sbjct: 352 TRAKVKDLHSRIRVAIHRINS 414
>CO978596
Length = 735
Score = 92.4 bits (228), Expect = 6e-19
Identities = 54/147 (36%), Positives = 79/147 (53%), Gaps = 3/147 (2%)
Frame = -1
Query: 515 GLAKMWKVMAECHQTQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNW 574
GL +MWK M ECH Q + + EA+IL I +RK+S S ++ LE EL NW
Sbjct: 735 GLTRMWKSMLECHHDQCEAIREARIL-GSIGSRKKSGDSHL------QATKQLEQELINW 577
Query: 575 RNTFESWITSQKSYIHALTGWLLRCMRCEPDASK---LICSPRRSSSTHPLFGLCIQWSR 631
F WI++QK Y+ AL WLL+C+ E + + + SP R + +F +C QWS+
Sbjct: 576 TFQFSGWISAQKGYVRALNNWLLKCLLYEXEETPDGIVPFSPGRIGAPQ-IFVICNQWSQ 400
Query: 632 RLDALEETAVLDSIDFFAAELGSFYAQ 658
LD + E V+DS+ F + + Q
Sbjct: 399 ALDRISEKEVVDSMHVFTMSVLQIWEQ 319
>TC211154 similar to GB|AAS99727.1|46518491|BT012583 At2g27090 {Arabidopsis
thaliana;} , partial (23%)
Length = 1116
Score = 78.6 bits (192), Expect = 9e-15
Identities = 60/232 (25%), Positives = 109/232 (46%), Gaps = 13/232 (5%)
Frame = +3
Query: 15 KICKDRKRFIKEAVEHRTQFANGHIAYIQSLKRVSAALLDYFEENEALEFSFDSFVTPPA 74
++C++RK+F+++A++ R A H++YIQSLK AL + E +E S + TP
Sbjct: 3 QLCRERKKFVRQALDGRCSLAAAHVSYIQSLKNTGTALRKFTEPEGPIEPSLYTTATPEQ 182
Query: 75 KNKASPAVISISKHSSPTTIE------FGP------NTTTLKVNYLRPSGNPPISVEERP 122
+ +S S S I+ F P +++ + N+++ S VEE+P
Sbjct: 183 PLALTERTLSFSSASVSHHIDAAEHENFSPTPSLPSSSSKFRANHMKHSTISSKKVEEKP 362
Query: 123 PSPEMVRVEMYSPMYQYGMDGFFGMQSQPMNASIFAYSPNNRPVIPPASPQSSQWDSFWN 182
P P + V Q+ + + A+ ++ P P QWD F+
Sbjct: 363 PVPVIGIVTSSGTT----------TQNTSVMSGTAAFEDSSLPAGTP------QWD-FFG 491
Query: 183 PFTSLDY-FGYPNGSSLEQIVMDDENRGLRKVREEEGIPDLEQEEMDDEQEG 233
F +D+ F + +G + Q + + ++ ++++REEEGIP+LE +E G
Sbjct: 492 LFHPIDHQFSFQDGKGMHQDIGNADD--IQRLREEEGIPELEDDEEKASSHG 641
>TC230368
Length = 895
Score = 68.6 bits (166), Expect = 1e-11
Identities = 58/219 (26%), Positives = 99/219 (44%), Gaps = 5/219 (2%)
Frame = +1
Query: 516 LAKMWKVMAECHQTQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNWR 575
L +MWK M ECH Q T+ A +S+ L + L E+ +
Sbjct: 115 LIRMWKAMLECHHAQYITISLAY--------HSRSTPGTLQGDALREIMTRLLEEVEFFG 270
Query: 576 NTFESWITSQKSYIHALTGWLLRCM--RCEPDASKLICSPRRSSSTHPLFGLCIQWSRRL 633
+F +WI S SY+ A+ WL C+ E S+ SPRR + P+F LC WS +
Sbjct: 271 LSFANWINSLTSYVEAVNAWLQNCILQPRERTKSRRPFSPRRVLAP-PIFVLCRDWSAGI 447
Query: 634 DALEETAVLDSIDFFAAELGSFYAQQLREDSAQNATVGSNVNMEMVEV-AKVGEEVMEVA 692
AL + +I F ++L Q + + Q ++VN M E +K EE + +
Sbjct: 448 KALPSEELSQAIRNFLSDL----HLQTEQHNDQLLKKQNSVNASMAETESKTNEENEDES 615
Query: 693 VKVLC--GGMSGAMSSMSEFAIDSAKGYNELVKQWENAK 729
+ C ++ + +++F+ S K Y ++ ++ E+A+
Sbjct: 616 TNLSCIHARLTKVLDRLTKFSEASLKMYEDIKQKSESAR 732
>AW277695
Length = 438
Score = 65.5 bits (158), Expect = 8e-11
Identities = 36/80 (45%), Positives = 47/80 (58%), Gaps = 9/80 (11%)
Frame = +1
Query: 1 MGCSQSKLDDEEAVKICKDRKRFIKEAVEHRTQFANGHIAYIQSLKRVSAALLDY----- 55
MGC+QSK+++EEAV CK+RKRF+K++V R FA H AY LK AAL D+
Sbjct: 37 MGCNQSKIENEEAVARCKERKRFMKDSVASRNAFAAAHSAYATCLKNTGAALGDFAHGEV 216
Query: 56 ----FEENEALEFSFDSFVT 71
F N+ S S+VT
Sbjct: 217 QNPQFHSNDNNTSSSSSYVT 276
>BM732799
Length = 421
Score = 58.2 bits (139), Expect = 1e-08
Identities = 36/141 (25%), Positives = 69/141 (48%), Gaps = 5/141 (3%)
Frame = +3
Query: 337 MAEVIKDLESQFTVVCNAANDVSALLEAKKAQYLSPSNELS-----ASKLLNPVALFRSS 391
+ +++ L+ F A +V+ +LEA + Y S + +++++ + RS
Sbjct: 24 LMQILNVLDDHFLKASEGAQEVTKMLEATRLHYHSNFADNRGHIDHSARVMRVITWNRSF 203
Query: 392 PSKIITNFSKTRDEVYDDPSEEQCVFSVSHQSTLDRLYAWEKKLYQEVKSGTRVRLAYEK 451
++N +D++ + E +H + LD+L AWEKKLY+EVK G ++ Y++
Sbjct: 204 RG--VSNGDAAKDDIDSEEYE-------THATVLDKLLAWEKKLYEEVKQGELMKFEYQR 356
Query: 452 KCLQLRNHDIKGEEPSSVDKT 472
K L +G S++KT
Sbjct: 357 KVAILNKQKKRGASAESLEKT 419
>BI700616
Length = 421
Score = 53.9 bits (128), Expect = 2e-07
Identities = 34/128 (26%), Positives = 56/128 (43%)
Frame = +1
Query: 469 VDKTRAAIRDLHTQITVSIHSVEAISRRIETLRDEELHSQLLELLQGLAKMWKVMAECHQ 528
+DKTR+++ L + + + + I + DEEL QL+ L GL +MW+ M E H+
Sbjct: 37 IDKTRSSVDKLESDLISLRQCISDTTSSILEMIDEELLPQLVALTAGLTQMWRTMHESHK 216
Query: 529 TQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNWRNTFESWITSQKSY 588
Q L +M + Y ++ ETE W N+F + Q+ Y
Sbjct: 217 AQALISQHLSNL------SDNHNMILNSEYH-HQATIQFETEASYWYNSFCKLVKFQREY 375
Query: 589 IHALTGWL 596
+ L W+
Sbjct: 376 VRTLYEWI 399
>AI899958
Length = 416
Score = 48.1 bits (113), Expect = 1e-05
Identities = 35/96 (36%), Positives = 47/96 (48%)
Frame = +2
Query: 135 PMYQYGMDGFFGMQSQPMNASIFAYSPNNRPVIPPASPQSSQWDSFWNPFTSLDYFGYPN 194
P Y Y D + + S +S ++ + P PP+ P+SS WD F N F + D YP
Sbjct: 65 PPYPYPYDPYSAVGS----SSPQLHAVSKPPPPPPSPPRSSTWD-FLNFFDNSDDKYYPQ 229
Query: 195 GSSLEQIVMDDENRGLRKVREEEGIPDLEQEEMDDE 230
+R R+VREEEGIPDLE E+ E
Sbjct: 230 ---THYPATATPSRDSREVREEEGIPDLEDEDYHHE 328
>CO985236
Length = 466
Score = 44.3 bits (103), Expect = 2e-04
Identities = 39/132 (29%), Positives = 63/132 (47%)
Frame = -1
Query: 208 RGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTKIDVNSSKEEVMVEDVDKHKEE 267
RGL ++R EEG+ + +++ +D E V E+R + V KE+ V+ V KE
Sbjct: 463 RGLVRIRHEEGVEEATEDKKEDGVE------EVNEDRKEDGVGEVKEDKEVDGVSCVKEV 302
Query: 268 EKEKGTDAETETAQEISDSKINGGECFQVSKSQSSGHMESSHKEMAIDTEEAKEKTPGFT 327
E+ D E + +++ D K G E ++ + + H+ S + +M DT E KE G
Sbjct: 301 EE----DGEVDGVKKVEDKK--GDELKEIEEDKKDDHI-SENDKMDEDT-EIKETMEG-- 152
Query: 328 VYVNRRPENMAE 339
PEN E
Sbjct: 151 -----EPENGKE 131
>TC226915 similar to UP|Q8LDZ7 (Q8LDZ7) Cinnamoyl CoA reductase-like protein,
partial (32%)
Length = 751
Score = 37.7 bits (86), Expect = 0.019
Identities = 34/140 (24%), Positives = 61/140 (43%), Gaps = 3/140 (2%)
Frame = +1
Query: 329 YVNRRPENMAEVIKDLESQFTVVCNAANDVSALLEAKKAQYLSPSNELSASKL-LNPVAL 387
+ NR P +K + ++ A DV+ L EA + + +N S + + V
Sbjct: 259 FCNRNPTATIAYLKGAQEMYSRRLLATVDVTKLAEAHASVFKEMNNNASGRYICFDHVID 438
Query: 388 FRSSPSKIITNFSKTRDEVYDDPSEEQCV--FSVSHQSTLDRLYAWEKKLYQEVKSGTRV 445
S K+ + +++ D S + F +S++ L RL + + Y E +
Sbjct: 439 THSEAEKLAKDIGMPEEKICGDASNNSIINRFELSNEK-LCRLMSRRLRCYSEY*MKIML 615
Query: 446 RLAYEKKCLQLRNHDIKGEE 465
LAY K CL+LR+ + K +E
Sbjct: 616 CLAYLKTCLRLRSLEEKIDE 675
>TC220482 similar to GB|AAS99727.1|46518491|BT012583 At2g27090 {Arabidopsis
thaliana;} , partial (13%)
Length = 550
Score = 37.4 bits (85), Expect = 0.024
Identities = 21/70 (30%), Positives = 36/70 (51%)
Frame = +1
Query: 516 LAKMWKVMAECHQTQKQTLDEAKILIAGIDARKQSSMSITDPYRLARSASNLETELRNWR 575
L++MW+VM ECH+ Q QT+ + + + ++ T + S LE+EL
Sbjct: 7 LSRMWEVMFECHKLQFQTM-------STVYNNSHAGIAATHSELRRQITSYLESELHYLS 165
Query: 576 NTFESWITSQ 585
++F WI +Q
Sbjct: 166 SSFTKWIGAQ 195
>TC224025 weakly similar to UP|Q6HXC4 (Q6HXC4) Conserved domain protein,
partial (15%)
Length = 614
Score = 37.4 bits (85), Expect = 0.024
Identities = 28/101 (27%), Positives = 48/101 (46%), Gaps = 1/101 (0%)
Frame = -1
Query: 188 DYFGYPNGSSLEQIVMDDE-NRGLRKVREEEGIPDLEQEEMDDEQEGCVVKRNVAEERTK 246
D G G ++ + D E ++G +++E+EG E+ D ++G EE +
Sbjct: 500 DCTGEKEGEKGKEELKDQEGDQGKEELKEQEGEKGKEELRELDAEKG-------KEELRE 342
Query: 247 IDVNSSKEEVMVEDVDKHKEEEKEKGTDAETETAQEISDSK 287
+D +KEE+ D +K KEE KE + E +E+ K
Sbjct: 341 LDAEKAKEELKELDGEKGKEELKELDGEKGKEELKELDGEK 219
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 34.7 bits (78), Expect = 0.16
Identities = 32/133 (24%), Positives = 60/133 (45%), Gaps = 11/133 (8%)
Frame = +2
Query: 204 DDENRGLRKVREEEGIPDL--EQEEMDDEQEGCVVKRNVA-EERTKIDVNSSKEEVMVED 260
DD++ G EE+ PD + ++ DDE+EG V + ++ D +S +E E+
Sbjct: 359 DDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGDVQRGGEPDDDDNDSDDDSDDDEDEDEE 538
Query: 261 VDKHKEEEKEKGTD--------AETETAQEISDSKINGGECFQVSKSQSSGHMESSHKEM 312
++ + EE++ GT+ AE E A + + NG E + + E+ K
Sbjct: 539 DEEEQGEEEDLGTEYLIRPLETAEEEEASSDFEPEENGEEEEEDVDDEDCEKAEAPPKRK 718
Query: 313 AIDTEEAKEKTPG 325
D +++ + G
Sbjct: 719 RSDKDDSDDDDGG 757
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,675,415
Number of Sequences: 63676
Number of extensions: 409145
Number of successful extensions: 2102
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 2022
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2071
length of query: 738
length of database: 12,639,632
effective HSP length: 104
effective length of query: 634
effective length of database: 6,017,328
effective search space: 3814985952
effective search space used: 3814985952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC125477.15


