

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
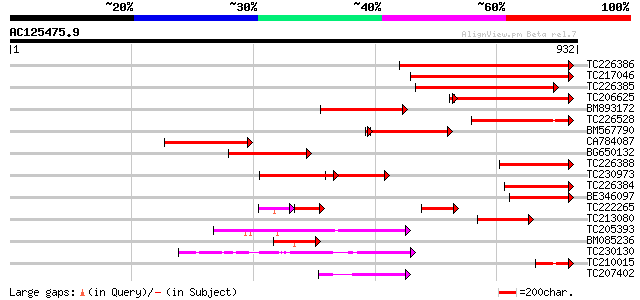
Query= AC125475.9 - phase: 0 /pseudo
(932 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226386 homologue to UP|Q9SAW3 (Q9SAW3) P-type H+-ATPase, parti... 526 e-149
TC217046 plasma membrane proton-ATPase 459 e-129
TC226385 homologue to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPa... 431 e-121
TC206625 homologue to UP|Q43106 (Q43106) H(+)-transporting ATPas... 315 3e-88
BM893172 PIR|T52414|T5 H+-exporting ATPase (EC 3.6.3.6) plasma ... 258 7e-69
TC226528 homologue to UP|Q7Y066 (Q7Y066) Plasma membrane H+-ATPa... 252 6e-67
BM567790 homologue to GP|20302435|em plasma membrane H+ ATPase {... 235 8e-65
CA784087 homologue to PIR|A43637|A43 H+-exporting ATPase (EC 3.6... 244 1e-64
BG650132 homologue to PIR|S52728|S52 H+-exporting ATPase (EC 3.6... 241 1e-63
TC226388 homologue to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPa... 233 3e-61
TC230973 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPa... 229 6e-60
TC226384 homologue to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPa... 214 2e-55
BE346097 similar to PIR|S52728|S52 H+-exporting ATPase (EC 3.6.3... 141 1e-33
TC222265 similar to UP|Q9SWH1 (Q9SWH1) Plasma membrane proton AT... 91 4e-33
TC213080 similar to UP|Q9ARG5 (Q9ARG5) Plasma membrane H+ ATPase... 134 2e-31
TC205393 UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase, partial... 100 4e-21
BM085236 similar to GP|12697490|emb p-type H+-ATPase {Sesbania r... 100 4e-21
TC230130 similar to UP|AHM7_ARATH (Q9SH30) Potential copper-tran... 92 1e-18
TC210015 homologue to UP|Q7Y066 (Q7Y066) Plasma membrane H+-ATPa... 84 3e-16
TC207402 similar to UP|Q941L1 (Q941L1) Copper-transporting P-typ... 70 4e-12
>TC226386 homologue to UP|Q9SAW3 (Q9SAW3) P-type H+-ATPase, partial (33%)
Length = 1352
Score = 526 bits (1354), Expect = e-149
Identities = 261/285 (91%), Positives = 269/285 (93%)
Frame = +2
Query: 642 AIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVV 701
AIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV
Sbjct: 11 AIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVK 190
Query: 702 PSPLPDSWKLKEIFATGIVLGGYLALMTVIFFWAMKENDFFPDKFGVRKLNHDEMMSALY 761
PSPLPDSWKL+EIFATGIVLG YLALMTVIFFWAMKE DFFPDKFGVR L HDEMMSALY
Sbjct: 191 PSPLPDSWKLQEIFATGIVLGSYLALMTVIFFWAMKETDFFPDKFGVRHLTHDEMMSALY 370
Query: 762 LQVSIVSQALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWG 821
LQVSIVSQALIFVTRSR WSF+ERPG L AF IAQLIATIIAVYA+WGFAKV+GIGWG
Sbjct: 371 LQVSIVSQALIFVTRSRSWSFIERPGMLFFCAFVIAQLIATIIAVYADWGFAKVKGIGWG 550
Query: 822 WAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHA 881
WAGVIWLYSIVFYIPLDVMKFA RYILSGKAW N+L+NKTAFTTKKDYGKEEREAQWAHA
Sbjct: 551 WAGVIWLYSIVFYIPLDVMKFATRYILSGKAWVNMLENKTAFTTKKDYGKEEREAQWAHA 730
Query: 882 QRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
QRTLHGLQPPE+SGIFNEK+SYRELSEIAEQAKRRAEVARL H
Sbjct: 731 QRTLHGLQPPETSGIFNEKNSYRELSEIAEQAKRRAEVARLRELH 865
>TC217046 plasma membrane proton-ATPase
Length = 1192
Score = 459 bits (1181), Expect = e-129
Identities = 219/269 (81%), Positives = 248/269 (91%), Gaps = 2/269 (0%)
Frame = +3
Query: 660 RIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGI 719
RIVFGFMFIALIWKFDF+PFMVLIIAILNDGTIMTISKDRV PSP+PDSWKL+EIFATGI
Sbjct: 3 RIVFGFMFIALIWKFDFAPFMVLIIAILNDGTIMTISKDRVKPSPMPDSWKLREIFATGI 182
Query: 720 VLGGYLALMTVIFFWAMKENDFFPDKFGVRKLNHD--EMMSALYLQVSIVSQALIFVTRS 777
VLG Y+ALMTV+FFW MK+ DFF DKFGVR + + EMM+ALYLQVSI+SQALIFVTRS
Sbjct: 183 VLGSYMALMTVVFFWIMKDTDFFSDKFGVRSIRNSPGEMMAALYLQVSIISQALIFVTRS 362
Query: 778 RGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPL 837
R WS++ERPG LL+ AF IAQL+AT +AVYANWGFA++QG+GWGWAGVIWLYS+V YIPL
Sbjct: 363 RSWSYVERPGLLLLSAFMIAQLVATFLAVYANWGFARIQGMGWGWAGVIWLYSLVTYIPL 542
Query: 838 DVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIF 897
D++KFAIRY+LSGKAW+NLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE++ +F
Sbjct: 543 DILKFAIRYVLSGKAWDNLLENKTAFTTKKDYGKEEREAQWATAQRTLHGLQPPETTNLF 722
Query: 898 NEKSSYRELSEIAEQAKRRAEVARLIPFH 926
N+K+SYRELSEIAEQAKRRAEVARL H
Sbjct: 723 NDKNSYRELSEIAEQAKRRAEVARLRELH 809
>TC226385 homologue to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPase, partial
(25%)
Length = 1109
Score = 431 bits (1108), Expect = e-121
Identities = 211/238 (88%), Positives = 222/238 (92%), Gaps = 2/238 (0%)
Frame = +1
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSPLPDSWKL EIFATG+VLGGYLA
Sbjct: 7 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLNEIFATGVVLGGYLA 186
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
LMTVIFFWA+KE FFPDKFGVR + N DEM +ALYLQVSIVSQALIFVTRSR WSF+E
Sbjct: 187 LMTVIFFWAIKETTFFPDKFGVRPIHDNPDEMTAALYLQVSIVSQALIFVTRSRSWSFIE 366
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LLV AF IAQLIAT+IAVYANWGFA++QGIGWGWAGVIWLYSIVFY PLD+MKFAI
Sbjct: 367 RPGLLLVTAFVIAQLIATVIAVYANWGFARIQGIGWGWAGVIWLYSIVFYFPLDIMKFAI 546
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSS 902
RYILSGKAWNNLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S IFNEKSS
Sbjct: 547 RYILSGKAWNNLLENKTAFTTKKDYGKEEREAQWALAQRTLHGLQPPETSNIFNEKSS 720
>TC206625 homologue to UP|Q43106 (Q43106) H(+)-transporting ATPase , partial
(24%)
Length = 1231
Score = 315 bits (807), Expect(2) = 3e-88
Identities = 155/198 (78%), Positives = 175/198 (88%), Gaps = 2/198 (1%)
Frame = +2
Query: 731 IFFWAMKENDFFPDKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSRGWSFLERPGA 788
IF K +FF +KFGVR L DEMM+ALYLQVSI+SQALIFVTRSR WSF+ERPG
Sbjct: 29 IFLGNXKIXNFFSNKFGVRPLRDSPDEMMAALYLQVSIISQALIFVTRSRSWSFVERPGL 208
Query: 789 LLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYIL 848
LL+ AFFIAQL+AT IAVYANW FA+++G+GWGWAGVIWLYS+V YIPLD++KFAIRYIL
Sbjct: 209 LLLGAFFIAQLVATFIAVYANWSFARIKGMGWGWAGVIWLYSVVTYIPLDLLKFAIRYIL 388
Query: 849 SGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSE 908
SGKAW+NLL+NKTAFTTKKDYGKEEREAQWA AQRTLHGLQPPE+S +FN+K+SYRELSE
Sbjct: 389 SGKAWDNLLENKTAFTTKKDYGKEEREAQWAAAQRTLHGLQPPETSNLFNDKNSYRELSE 568
Query: 909 IAEQAKRRAEVARLIPFH 926
IAEQAKRRAEVARL H
Sbjct: 569 IAEQAKRRAEVARLRELH 622
Score = 30.0 bits (66), Expect(2) = 3e-88
Identities = 11/13 (84%), Positives = 13/13 (99%)
Frame = +3
Query: 724 YLALMTVIFFWAM 736
Y+ALMTV+FFWAM
Sbjct: 6 YMALMTVVFFWAM 44
>BM893172 PIR|T52414|T5 H+-exporting ATPase (EC 3.6.3.6) plasma membrane
[imported] - Prunus persica, partial (14%)
Length = 427
Score = 258 bits (660), Expect = 7e-69
Identities = 129/142 (90%), Positives = 136/142 (94%)
Frame = -2
Query: 512 VKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPVEELIEKADGFAGVFP 571
VKMITGDQLAI KETGRRLGMGTNMYPS+ LLGQDKD +I ALP++ELIEKADGFAGVFP
Sbjct: 426 VKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQDKDESIVALPIDELIEKADGFAGVFP 247
Query: 572 EHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDIVLTEPGLS 631
EHKYEIV++LQ RKHICGMTGDGVNDAPALK+ADIGIAVADATDAAR ASDIVLTEPGLS
Sbjct: 246 EHKYEIVKRLQARKHICGMTGDGVNDAPALKKADIGIAVADATDAARSASDIVLTEPGLS 67
Query: 632 VIISAVLTSRAIFQRMKNYTIY 653
VIISAVLTSRAIFQRMKNYTIY
Sbjct: 66 VIISAVLTSRAIFQRMKNYTIY 1
>TC226528 homologue to UP|Q7Y066 (Q7Y066) Plasma membrane H+-ATPase, partial
(20%)
Length = 962
Score = 252 bits (643), Expect = 6e-67
Identities = 116/168 (69%), Positives = 145/168 (86%)
Frame = +3
Query: 759 ALYLQVSIVSQALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGI 818
A+YLQVS +SQALIFVTRSRGWS++ERPG LLV AF IAQLIAT+IAVYANW FA ++GI
Sbjct: 3 AIYLQVSTISQALIFVTRSRGWSYVERPGILLVTAFVIAQLIATLIAVYANWSFAAIEGI 182
Query: 819 GWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQW 878
GWGWAGVIWLY+I+FYIPLD +KF IRY LSG+AW +++ + AFT +KD+GKE+RE QW
Sbjct: 183 GWGWAGVIWLYNIIFYIPLDPIKFLIRYALSGRAWELVIEQRIAFTRQKDFGKEQRELQW 362
Query: 879 AHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
AHAQRTLHGLQPP++ +F E++ + EL+++AE+AKRRAE+ARL H
Sbjct: 363 AHAQRTLHGLQPPDTK-MFTERTHFNELNQMAEEAKRRAEIARLRELH 503
>BM567790 homologue to GP|20302435|em plasma membrane H+ ATPase {Oryza sativa
(japonica cultivar-group)}, partial (15%)
Length = 436
Score = 235 bits (600), Expect(2) = 8e-65
Identities = 120/135 (88%), Positives = 125/135 (91%)
Frame = +3
Query: 594 GVNDAPALKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIY 653
G+ DAPALK+ADIGIAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIY
Sbjct: 30 GLTDAPALKKADIGIAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIY 209
Query: 654 AVSITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKE 713
AVSITIRIV GFM +ALIW FDF PFMVLIIAILNDGTIMTISKDRV PSP PDSWKL E
Sbjct: 210 AVSITIRIVLGFMLLALIWHFDFPPFMVLIIAILNDGTIMTISKDRVKPSPYPDSWKLAE 389
Query: 714 IFATGIVLGGYLALM 728
IF TGI+LGGYLA+M
Sbjct: 390 IFTTGIILGGYLAMM 434
Score = 31.6 bits (70), Expect(2) = 8e-65
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +2
Query: 585 KHICGMTGDGVN 596
KHICGMTGDGVN
Sbjct: 2 KHICGMTGDGVN 37
>CA784087 homologue to PIR|A43637|A43 H+-exporting ATPase (EC 3.6.3.6) -
curled-leaved tobacco, partial (15%)
Length = 435
Score = 244 bits (624), Expect = 1e-64
Identities = 124/145 (85%), Positives = 137/145 (93%)
Frame = -1
Query: 255 SIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGA 314
SIAVGM+ EIIVMYPIQHR+YR GIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRL+QQGA
Sbjct: 435 SIAVGMIVEIIVMYPIQHREYRPGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLAQQGA 256
Query: 315 ITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVE 374
ITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIE+F KGVD + V+ +AARA+R+E
Sbjct: 255 ITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEIFAKGVDVDTVVQMAARAARLE 76
Query: 375 NQDAIDAAIVGTLADPKEARAGVRE 399
NQDAIDA+IVG L DPKEARAG++E
Sbjct: 75 NQDAIDASIVGMLGDPKEARAGIQE 1
>BG650132 homologue to PIR|S52728|S52 H+-exporting ATPase (EC 3.6.3.6) -
kidney bean, partial (14%)
Length = 412
Score = 241 bits (615), Expect = 1e-63
Identities = 113/137 (82%), Positives = 126/137 (91%)
Frame = +2
Query: 360 KEHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDG 419
+E+V+LLAARASR ENQDAIDAAIVG LADPKEAR+GVRE+HFLPFNPVDKRTALTYID
Sbjct: 2 REYVILLAARASRTENQDAIDAAIVGMLADPKEARSGVREVHFLPFNPVDKRTALTYIDS 181
Query: 420 NGNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPG 479
+GNWHRASKGAPEQI+ LC +ED +R +HA+IDKFAERGLRSL VARQEVPEK+K+SPG
Sbjct: 182 DGNWHRASKGAPEQIITLCNCKEDVRRKVHAVIDKFAERGLRSLGVARQEVPEKSKDSPG 361
Query: 480 APWQFVGLLSLFDPPRH 496
PWQFVGLL LFDPPRH
Sbjct: 362 GPWQFVGLLPLFDPPRH 412
>TC226388 homologue to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPase, partial
(16%)
Length = 619
Score = 233 bits (594), Expect = 3e-61
Identities = 110/122 (90%), Positives = 116/122 (94%)
Frame = +3
Query: 805 AVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFT 864
AVYANWGFA++QGIGWGWAGVIWLYSIVFY PLD+MKFAIRYILSGKAWNNLL+NKTAFT
Sbjct: 3 AVYANWGFARIQGIGWGWAGVIWLYSIVFYFPLDLMKFAIRYILSGKAWNNLLENKTAFT 182
Query: 865 TKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIP 924
TKKDYGKEEREAQWA AQRTLHGLQPPE+S IFNEKSSYREL+EIAEQAKRRAEVARL
Sbjct: 183 TKKDYGKEEREAQWALAQRTLHGLQPPETSNIFNEKSSYRELTEIAEQAKRRAEVARLRE 362
Query: 925 FH 926
H
Sbjct: 363 LH 368
>TC230973 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPase, partial
(22%)
Length = 654
Score = 229 bits (583), Expect = 6e-60
Identities = 108/130 (83%), Positives = 119/130 (91%)
Frame = +2
Query: 411 RTALTYIDGNGNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEV 470
RTALTYID +GNWHR+SKGAPEQI++LC +ED ++ +H IDKFAERGLRSL VARQEV
Sbjct: 14 RTALTYIDSDGNWHRSSKGAPEQILNLCNCKEDVRKRVHGTIDKFAERGLRSLGVARQEV 193
Query: 471 PEKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRL 530
PEK K+SPGAPWQFVGLL LFDPPRHDSAETI RAL+LGVNVKMITGDQLAIAKETGRRL
Sbjct: 194 PEKNKDSPGAPWQFVGLLPLFDPPRHDSAETITRALNLGVNVKMITGDQLAIAKETGRRL 373
Query: 531 GMGTNMYPSA 540
GMGTNMYPS+
Sbjct: 374 GMGTNMYPSS 403
Score = 152 bits (383), Expect = 9e-37
Identities = 76/104 (73%), Positives = 86/104 (82%)
Frame = +3
Query: 520 LAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVR 579
L + K+ LG +LLGQ KDA ++A+PV+ELIEKADGFAGVFPEHKYEIV+
Sbjct: 342 LPLPKKLAEGLGWEQTCTHLPSLLGQSKDAAVSAVPVDELIEKADGFAGVFPEHKYEIVK 521
Query: 580 KLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDI 623
+LQERKHICGMTGDGVNDAPALK+ADIGIAVADATDAAR ASDI
Sbjct: 522 RLQERKHICGMTGDGVNDAPALKKADIGIAVADATDAARSASDI 653
>TC226384 homologue to UP|Q7Y067 (Q7Y067) Plasma membrane H+-ATPase, partial
(15%)
Length = 754
Score = 214 bits (544), Expect = 2e-55
Identities = 101/113 (89%), Positives = 107/113 (94%)
Frame = +1
Query: 814 KVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEE 873
++QGIGWGWAGVIWLYSIVFY PLD+MKFAIRYILSGKAWNNLL+NKTAFTTKKDYGKEE
Sbjct: 1 RIQGIGWGWAGVIWLYSIVFYFPLDIMKFAIRYILSGKAWNNLLENKTAFTTKKDYGKEE 180
Query: 874 REAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
REAQWA AQRTLHGLQPPE+S IFNEKSSYREL+EIAEQAKRRAEVARL H
Sbjct: 181 REAQWALAQRTLHGLQPPETSNIFNEKSSYRELTEIAEQAKRRAEVARLRELH 339
>BE346097 similar to PIR|S52728|S52 H+-exporting ATPase (EC 3.6.3.6) - kidney
bean, partial (13%)
Length = 394
Score = 141 bits (356), Expect = 1e-33
Identities = 69/105 (65%), Positives = 82/105 (77%)
Frame = +2
Query: 822 WAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHA 881
W GV+WL +V YI LD++K+AIRY LSGKAW+NLLDNKTA TTK DYG EE EA WA A
Sbjct: 2 WVGVMWLLCLVTYIRLDLLKYAIRYFLSGKAWDNLLDNKTALTTKNDYG*EETEAHWATA 181
Query: 882 QRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLIPFH 926
QRTLHGLQ PE++ +FN+ +S EL +IA QA+RRA VARL H
Sbjct: 182 QRTLHGLQTPETTNLFNDNNSCWELLQIAYQAQRRA*VARLRELH 316
>TC222265 similar to UP|Q9SWH1 (Q9SWH1) Plasma membrane proton ATPase,
partial (16%)
Length = 792
Score = 90.5 bits (223), Expect(2) = 4e-33
Identities = 42/49 (85%), Positives = 45/49 (91%)
Frame = +2
Query: 469 EVPEKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITG 517
EVPE TK+SPG PW+FVGLL LFDPPRHDSAETIRRAL LGV+VKMITG
Sbjct: 353 EVPEGTKDSPGGPWEFVGLLPLFDPPRHDSAETIRRALDLGVSVKMITG 499
Score = 85.5 bits (210), Expect = 1e-16
Identities = 44/62 (70%), Positives = 49/62 (78%)
Frame = +2
Query: 677 SPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALMTVIFFWAM 736
S F+V + + GTIMTISKDRV PSPLPDSWKL EIF TGIVLG YLALMTVIFF+ +
Sbjct: 602 SLFLVP*LVSILVGTIMTISKDRVKPSPLPDSWKLSEIFTTGIVLGSYLALMTVIFFYIV 781
Query: 737 KE 738
E
Sbjct: 782 VE 787
Score = 70.5 bits (171), Expect(2) = 4e-33
Identities = 41/90 (45%), Positives = 49/90 (53%), Gaps = 30/90 (33%)
Frame = +1
Query: 409 DKRTALTYIDGNGNWHRASKGAPEQ------------------------------IMDLC 438
DKRTALTY+D G HR SKGAPEQ I++L
Sbjct: 1 DKRTALTYLDAAGKMHRVSKGAPEQVEHNPL*L*RTILPTRVTFV*SLFNSSTV*ILNLA 180
Query: 439 KLREDTKRNIHAIIDKFAERGLRSLAVARQ 468
+ + ++ +HAIIDKFAERGLRSLAVARQ
Sbjct: 181 HNKSEIQQRVHAIIDKFAERGLRSLAVARQ 270
>TC213080 similar to UP|Q9ARG5 (Q9ARG5) Plasma membrane H+ ATPase, partial
(10%)
Length = 427
Score = 134 bits (336), Expect = 2e-31
Identities = 58/92 (63%), Positives = 75/92 (81%)
Frame = +2
Query: 769 QALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWL 828
+ALIFVTRSR WS+ ERPG +L +AF AQL+AT+IAVYA+W FA++ G+GW WAGVIW+
Sbjct: 14 EALIFVTRSRSWSYFERPGIMLCVAFICAQLVATVIAVYAHWDFARINGVGWRWAGVIWI 193
Query: 829 YSIVFYIPLDVMKFAIRYILSGKAWNNLLDNK 860
YSI+ YIPLD++KF IR L+G A +N+ NK
Sbjct: 194 YSIITYIPLDILKFLIRMGLTGSAGDNMHQNK 289
>TC205393 UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase, partial (55%)
Length = 2009
Score = 100 bits (248), Expect = 4e-21
Identities = 87/371 (23%), Positives = 158/371 (42%), Gaps = 46/371 (12%)
Frame = +2
Query: 335 KTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQDAIDAAIVGT-------- 386
KTGTLT N ++V K + K V + L + + +++ T
Sbjct: 2 KTGTLTTNHMTVVKTCFCMNSKEVSNNNASSLCSELPEPAVKLLLESIFNNTGGEVVVNQ 181
Query: 387 ------LADPKEA----------------RAGVREIHFLPFNPVDKRTALTYIDGNGNWH 424
L P EA + + + PFN K+ ++ G
Sbjct: 182 NGKREILGTPTEAAILEFGLSLGGDFQGEKQACKLVKVEPFNSTKKKMSVVVELPGGGLR 361
Query: 425 RASKGAPEQIMDLCK-----------LREDTKRNIHAIIDKFAERGLRSLAVARQEVPEK 473
KGA E I+ C L E++ ++ A I++FA LR+L +A E+
Sbjct: 362 AHCKGASEIILAACDKVLNSNGEVVPLDEESTSHLKATINQFASEALRTLCLAYVELENG 541
Query: 474 TKESPGAP---WQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRL 530
P + +G++ + DP R E++ G+ V+M+TGD + AK R
Sbjct: 542 FSPEDPIPVSGYTCIGVIGIKDPVRPGVKESVAM*RSAGITVRMVTGDNINTAKAIAREC 721
Query: 531 GMGTNMYPSATLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQER-KHICG 589
G+ T+ + + + + ELI K A P K+ +V+ L+ +
Sbjct: 722 GILTD---DGIAIEGPEFREKSQKELLELIPKIQVMARSSPLDKHTLVKHLRTTFGEVVA 892
Query: 590 MTGDGVNDAPALKRADIGIAVADA-TDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMK 648
+TGDG NDAPAL ADIG+A+ A T+ A+ ++D+++ + S I++ R+++ ++
Sbjct: 893 VTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQ 1072
Query: 649 NYTIYAVSITI 659
+ + +++ +
Sbjct: 1073KFVQFQLTVNV 1105
>BM085236 similar to GP|12697490|emb p-type H+-ATPase {Sesbania rostrata},
partial (31%)
Length = 407
Score = 100 bits (248), Expect = 4e-21
Identities = 53/110 (48%), Positives = 67/110 (60%), Gaps = 32/110 (29%)
Frame = +3
Query: 434 IMDLCKLREDTKRNIHAIIDKFAERGLRSLAVA--------------------------- 466
I++L + + + +R +H++IDKFA+RGLRSLAVA
Sbjct: 78 ILNLARNKSEIERRVHSVIDKFADRGLRSLAVAYQVHMHLCSFFCGGDSKFKLQCMSLLT 257
Query: 467 -----RQEVPEKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVN 511
QEVP+ KES G PWQF+GLL LFDPPRHDSA+TIRRAL+LGVN
Sbjct: 258 AFIIISQEVPDGKKESQGGPWQFIGLLPLFDPPRHDSAQTIRRALNLGVN 407
>TC230130 similar to UP|AHM7_ARATH (Q9SH30) Potential copper-transporting
ATPase 3 , partial (43%)
Length = 1409
Score = 92.0 bits (227), Expect = 1e-18
Identities = 104/393 (26%), Positives = 168/393 (42%), Gaps = 3/393 (0%)
Frame = +1
Query: 278 GIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTG 337
GI +++ + +A PT + V +G+ QG + K A+E +D + DKTG
Sbjct: 118 GISVMVIACPCALGLATPTAVMVGTGVGA----SQGVLIKGGQALESAHKVDCIVFDKTG 285
Query: 338 TLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGV 397
TLT+ K + + E+ K V +E L+AA + V ++ + A+V ++
Sbjct: 286 TLTVGKPVIVRT--ELLTKMVLQEFYELVAA--AEVNSEHPLAKAVVEYAKRFRDEE--- 444
Query: 398 REIHFLPFNPVDKRTALTYIDGNGNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAE 457
NP A ++ G+ +AS E I+ L D NI AI D
Sbjct: 445 --------NP-SWPEARDFVSITGHGVKASVHNKEIIVGNKSLFAD--HNI-AIPDD--- 579
Query: 458 RGLRSLAVARQEVPEKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITG 517
+A E +T + G+L++ DP + + E I + + M+TG
Sbjct: 580 ---AEYILAEAEKMAQTGIVVSINGKVAGVLAVSDPLKPGAQEVISILKSMKIKSIMVTG 750
Query: 518 DQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEI 577
D A R +G +E +I +A P+ K E
Sbjct: 751 DNFGTASSIAREVG------------------------IENVIAEAK------PDQKAEK 840
Query: 578 VRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDIVLTEPGLSVIISAV 637
V+ LQ + M GDG+ND+PAL AD+G+A+ TD A A+DIVL + L +I+A+
Sbjct: 841 VKDLQASGYTVAMVGDGINDSPALVAADVGMAIGAGTDIAIEAADIVLMKSNLEDVITAI 1020
Query: 638 LTSRAIFQRMKNYTIYAVS---ITIRIVFGFMF 667
SR F R++ +A+ + I I G +F
Sbjct: 1021DLSRKTFSRIRLNYFWALGYNLLGIPIAAGALF 1119
>TC210015 homologue to UP|Q7Y066 (Q7Y066) Plasma membrane H+-ATPase, partial
(9%)
Length = 526
Score = 84.0 bits (206), Expect = 3e-16
Identities = 39/63 (61%), Positives = 51/63 (80%)
Frame = +2
Query: 864 TTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVARLI 923
T +KD+GKE+RE QWAHAQRTLHGLQP ++ FNE++ EL+++AE+AKRRAE+ARL
Sbjct: 2 TRQKDFGKEQRELQWAHAQRTLHGLQPADTK--FNERTHVNELNQMAEEAKRRAEIARLR 175
Query: 924 PFH 926
H
Sbjct: 176 ELH 184
>TC207402 similar to UP|Q941L1 (Q941L1) Copper-transporting P-type ATPase,
partial (19%)
Length = 721
Score = 70.1 bits (170), Expect = 4e-12
Identities = 44/152 (28%), Positives = 70/152 (45%)
Frame = +1
Query: 508 LGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPVEELIEKADGFA 567
+GV M+TGD A+ + +G+ D A
Sbjct: 31 MGVTPVMVTGDNWRTARAVAKEVGI------------------------------QDVRA 120
Query: 568 GVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDIVLTE 627
V P K ++VR Q+ I M GDG+ND+PAL AD+G+A+ TD A A++ VL
Sbjct: 121 EVMPAGKADVVRSFQKDGSIAAMVGDGINDSPALAAADVGMAIGAGTDIAIEAAEYVLMR 300
Query: 628 PGLSVIISAVLTSRAIFQRMKNYTIYAVSITI 659
L +I+A+ SR F R++ ++A++ +
Sbjct: 301 NNLEDVITAIDLSRKTFSRIRLNYVFAMAYNV 396
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,804,669
Number of Sequences: 63676
Number of extensions: 441943
Number of successful extensions: 2217
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 2179
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2207
length of query: 932
length of database: 12,639,632
effective HSP length: 106
effective length of query: 826
effective length of database: 5,889,976
effective search space: 4865120176
effective search space used: 4865120176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC125475.9


