

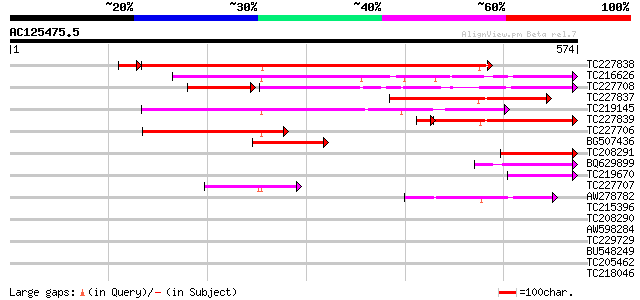
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.5 + phase: 0 /pseudo
(574 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227838 similar to UP|VIL4_ARATH (O65570) Villin 4, partial (38%) 520 e-150
TC216626 similar to UP|VIL2_ARATH (O81644) Villin 2, partial (36%) 269 2e-72
TC227708 weakly similar to UP|Q9LVC6 (Q9LVC6) Villin, partial (17%) 214 8e-67
TC227837 similar to UP|VIL4_ARATH (O65570) Villin 4, partial (12%) 239 2e-63
TC219145 similar to UP|VIL2_ARATH (O81644) Villin 2, partial (35%) 230 1e-60
TC227839 similar to UP|VIL4_ARATH (O65570) Villin 4, partial (12%) 199 2e-56
TC227706 similar to UP|VIL1_ARATH (O81643) Villin 1, partial (11%) 152 4e-37
BG507436 similar to GP|8777365|dbj| villin {Arabidopsis thaliana... 115 4e-26
TC208291 similar to UP|Q7Y0V3 (Q7Y0V3) Actin filament bundling p... 114 1e-25
BQ629899 weakly similar to PIR|T50669|T506 villin 2 [imported] -... 70 2e-12
TC219670 weakly similar to UP|VIL2_ARATH (O81644) Villin 2, part... 65 6e-11
TC227707 similar to UP|VIL1_ARATH (O81643) Villin 1, partial (5%) 63 4e-10
AW278782 similar to PIR|T50669|T506 villin 2 [imported] - Arabid... 57 2e-08
TC215396 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kD chloroplast ... 38 0.014
TC208290 35 0.12
AW598284 32 0.77
TC229729 similar to GB|AAO63276.1|28950705|BT005212 At2g42260 {A... 32 0.77
BU548249 31 1.3
TC205462 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MS... 31 1.7
TC218046 weakly similar to PIR|T49196|T49196 nematode resistance... 31 1.7
>TC227838 similar to UP|VIL4_ARATH (O65570) Villin 4, partial (38%)
Length = 1235
Score = 520 bits (1339), Expect(2) = e-150
Identities = 266/367 (72%), Positives = 309/367 (83%), Gaps = 12/367 (3%)
Frame = +2
Query: 134 DETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDD 193
D+TY E+GVALFRIQGSGP++MQAIQV ASSLNSSYCYIL + VFT +GN T++++
Sbjct: 137 DDTYNENGVALFRIQGSGPDNMQAIQVEPVASSLNSSYCYILHNGPAVFTCFGNSTSAEN 316
Query: 194 QELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFS 253
QEL ERMLDLIKP+LQ +PQ+EG+E+EQFW+ LG K+EY SQKI+RE E+DPHLFSC+FS
Sbjct: 317 QELVERMLDLIKPNLQSKPQREGSESEQFWDFLGGKSEYPSQKILREPESDPHLFSCHFS 496
Query: 254 E------EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFL 307
+ E++NFSQDDLMTEDIFILDCHS+IFVWVGQQVD K R+QAL IGEKFLE DFL
Sbjct: 497 KGNLKVTEVYNFSQDDLMTEDIFILDCHSEIFVWVGQQVDSKSRMQALTIGEKFLEHDFL 676
Query: 308 LETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKR 367
LE +S AP+Y+VMEGSEPPFFTRFFKWDSAKS+MLGNS+QRKL I+K+GG P L KPKR
Sbjct: 677 LEKLSHVAPVYVVMEGSEPPFFTRFFKWDSAKSSMLGNSFQRKLTIVKSGGAPVLDKPKR 856
Query: 368 RASVSYGGRSGGLPEKSQR--SRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPP 425
R VSYGGRS +P+KS + SRSMSVSPDRVRVRGRSPAFNALAA FEN N RNLSTPP
Sbjct: 857 RTPVSYGGRSSSVPDKSSQRSSRSMSVSPDRVRVRGRSPAFNALAANFENPNARNLSTPP 1036
Query: 426 PMIRKLYPKSKTPDLATLAPKSSAISHLTSTFE-PPSAREKLIPRSLKDT---SKSNPET 481
P+IRKLYPKS T D A LAPKSSAI+ L+S+FE PPSARE +IP+S+K + KSNPE
Sbjct: 1037PVIRKLYPKSVTTDSAILAPKSSAIAALSSSFEQPPSARETMIPKSIKVSPVMPKSNPEK 1216
Query: 482 NSDNENS 488
N D ENS
Sbjct: 1217N-DKENS 1234
Score = 30.0 bits (66), Expect(2) = e-150
Identities = 14/23 (60%), Positives = 16/23 (68%)
Frame = +1
Query: 111 GSYL*RQ*NNSILFHPPKLDCF* 133
GSY+*RQ NSI F+P CF*
Sbjct: 16 GSYI*RQRTNSISFYPSPFHCF* 84
>TC216626 similar to UP|VIL2_ARATH (O81644) Villin 2, partial (36%)
Length = 1777
Score = 269 bits (688), Expect = 2e-72
Identities = 158/447 (35%), Positives = 243/447 (54%), Gaps = 38/447 (8%)
Frame = +3
Query: 166 SLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIKPDLQCRPQKEGAETEQFWEL 225
SLNS+ C++LQS S +FTW+GN + + Q+LA ++ D ++P + KEG E+ FW
Sbjct: 3 SLNSTECFVLQSGSTIFTWHGNQCSFEQQQLAAKVADFLRPGATLKHAKEGTESSAFWSA 182
Query: 226 LGVKTEYSSQKIVREAENDPHLFSCNFS------EEIHNFSQDDLMTEDIFILDCHSQIF 279
LG K Y+S+K+V E DPHLF+ +F+ EE++NFSQDDL+ EDI ILD H+++F
Sbjct: 183 LGGKQSYTSKKVVNEVVRDPHLFTLSFNKGKFNVEEVYNFSQDDLLPEDILILDTHAEVF 362
Query: 280 VWVGQQVDPKRRVQALPIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAK 339
+W+G V+PK + A IG+K+++ LE +S P+Y V EG+EP FFT +F WD AK
Sbjct: 363 IWIGHSVEPKEKRNAFEIGQKYIDLVASLEGLSPHVPLYKVTEGNEPCFFTTYFSWDHAK 542
Query: 340 SAMLGNSYQRKLAIM----------KNGGTPPLVKPKRRASVSYGGRSGGLPEKSQRSRS 389
+ ++GNS+Q+K++++ NG +P + + A + G EK+
Sbjct: 543 AMVMGNSFQKKVSLLFGLGHAVEEKLNGSSPGGPRQRAEALAALSNAFGSSSEKAS---- 710
Query: 390 MSVSPDRVR------VRGRSPAFNALAATFENSNVRNLSTPPPMIR-------------- 429
++ DR+ R R+ A AL + F +S+ TP P R
Sbjct: 711 -GLAQDRLNGLGQGGPRQRAEALAALNSAFNSSSGTKTFTPRPSGRGQGSQRAAAVAALS 887
Query: 430 --KLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSDNEN 487
+ K K+PD + +A + S I+ ++T + E K+T + PE
Sbjct: 888 QVLMAEKKKSPDGSPVASR-SPITEGSATETKSDSSEVEEVAEAKETEELPPE------- 1043
Query: 488 STGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEFQ 547
TGS + QE+ EG N+G ++ YE +KT S +P +D+ +REAYLS +EF
Sbjct: 1044-TGSNGDLELKQENAEEG----NDGQRMFSYEQLKTKSGHNVPGVDLKRREAYLSEDEFN 1208
Query: 548 ERLGMTRSEFYKLPKWKQNKLKMAVQL 574
GM + FYKLP+WKQ+ LK +L
Sbjct: 1209TVFGMAKEAFYKLPRWKQDMLKKKYEL 1289
>TC227708 weakly similar to UP|Q9LVC6 (Q9LVC6) Villin, partial (17%)
Length = 1262
Score = 214 bits (544), Expect(2) = 8e-67
Identities = 128/321 (39%), Positives = 181/321 (55%)
Frame = +1
Query: 254 EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLETISC 313
+EI+N++QDDL+TEDI +LDC +I+VWVG K + +AL +G KFLE D L+E +S
Sbjct: 232 KEIYNYTQDDLITEDILLLDCQREIYVWVGLHSAIKSKQEALNLGLKFLEMDVLVEGLSM 411
Query: 314 SAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKRRASVSY 373
+ PIYIV EG EPPFFTRFF WD +K + GNS++RKLAI+K G P ++ R +
Sbjct: 412 NIPIYIVTEGHEPPFFTRFFSWDHSKENIFGNSFERKLAILK--GKPKSLEGHNRTPLKA 585
Query: 374 GGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYP 433
R P S+SV + R R SP ++ + S R+LS+ P+++KL+
Sbjct: 586 NSR----PSTPDGHGSISVFSNG-RGRSSSPIPSSAGSDLRQSGDRSLSSSTPVVKKLFE 750
Query: 434 KSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSDNENSTGSRE 493
S + +SSA K+ P++ S + S E
Sbjct: 751 GSPS--------QSSA-------------------------GKTMPQSGSPATELSSSDE 831
Query: 494 ESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEFQERLGMT 553
+ Q+D N D E +YPYE ++ S +P+ ID+TKRE YLS EEF+E+ GM
Sbjct: 832 TASFPQKDRN----VDGENTAIYPYERLRVVSANPVTGIDLTKREVYLSNEEFREKFGMP 999
Query: 554 RSEFYKLPKWKQNKLKMAVQL 574
+S FYKLP+WKQNKLKM++ L
Sbjct: 1000KSAFYKLPRWKQNKLKMSLDL 1062
Score = 58.9 bits (141), Expect(2) = 8e-67
Identities = 26/69 (37%), Positives = 43/69 (61%)
Frame = +3
Query: 181 VFTWYGNLTNSDDQELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVRE 240
++TW G+L+++ D L +RM++L+ P +EG E + FW+ LG K EY K ++
Sbjct: 6 IYTWIGSLSSARDHNLLDRMVELLNPTWLPVSVREGNEPDIFWDALGGKAEYPKGKEIQG 185
Query: 241 AENDPHLFS 249
+DPHLF+
Sbjct: 186 FIDDPHLFA 212
>TC227837 similar to UP|VIL4_ARATH (O65570) Villin 4, partial (12%)
Length = 499
Score = 239 bits (610), Expect = 2e-63
Identities = 130/167 (77%), Positives = 136/167 (80%), Gaps = 3/167 (1%)
Frame = +2
Query: 385 QRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLA 444
QRSRSMSVSPDRVRVRGRSPAFNALAA FE+SN RNLSTPPPMIRKLYPKS D A L
Sbjct: 2 QRSRSMSVSPDRVRVRGRSPAFNALAANFESSNARNLSTPPPMIRKLYPKSVAKDTAQLV 181
Query: 445 PKSSAISHLTSTFEPPSAREKLIPRSLKD---TSKSNPETNSDNENSTGSREESLTIQED 501
PKSSAI+HLTS+FEP SA E LIP+S K T KSNPET SD E S SR ESLTIQED
Sbjct: 182 PKSSAIAHLTSSFEPFSALENLIPQSQKANSVTPKSNPET-SDKEGSMSSRIESLTIQED 358
Query: 502 VNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEFQE 548
V EGE ED+EGLPVYPYE V T STDP+ DIDVTKREAYLS EFQE
Sbjct: 359 VKEGEAEDDEGLPVYPYERVNTASTDPVEDIDVTKREAYLSSAEFQE 499
>TC219145 similar to UP|VIL2_ARATH (O81644) Villin 2, partial (35%)
Length = 1261
Score = 230 bits (587), Expect = 1e-60
Identities = 132/392 (33%), Positives = 212/392 (53%), Gaps = 19/392 (4%)
Frame = +2
Query: 134 DETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDD 193
DETY + VA RI G+ + + +QV++ A+ LNS+ C++LQS S VFTW+GN + +
Sbjct: 116 DETYTAESVAFIRISGTSTHNNKVVQVDAVAALLNSTECFVLQSGSAVFTWHGNQCSLEQ 295
Query: 194 QELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFS 253
Q+LA ++ + ++P + + KEG ET FW LG K Y+S+ + + DPHLF+ +F+
Sbjct: 296 QQLAAKVAEFLRPGVSLKLAKEGTETSTFWFALGGKQSYTSKNVTNDIVRDPHLFTLSFN 475
Query: 254 ------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFL 307
EE++NFSQDDL+TEDI ILD H+++FVW+GQ VDPK + +A I +K++++
Sbjct: 476 RGKLQVEEVYNFSQDDLLTEDILILDTHTEVFVWIGQCVDPKEKQKAFEIAQKYIDKAAS 655
Query: 308 LETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKR 367
LE +S P+Y V EG+EP FFT +F WD K+ + GNS+Q+K+ ++ G P V+ K
Sbjct: 656 LEGLSPHVPLYKVTEGNEPCFFTTYFSWDHTKAMVPGNSFQKKVTLLFGIGHP--VEEKS 829
Query: 368 RASVSYGGRSGGLPEKSQRSRSMSVSPD------------RVRVRGRSPAFNALAATFEN 415
S GG + + + + SP+ R R R+ A AL + F +
Sbjct: 830 NGSSQGGGPRQRAEPLAALNNAFNSSPEATSSADKSNGLSRGGPRQRAEALAALNSAFNS 1009
Query: 416 SNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTS 475
S+ + TP P R + +++A++ L+S + TS
Sbjct: 1010SSGTKVYTPKPSGR-----------GQGSQRAAAVTALSSVLTAEKKKTAPETSPADSTS 1156
Query: 476 KSNPETNSDNENSTGSREESLTIQ-EDVNEGE 506
+N D ++ + E+ + + DV E E
Sbjct: 1157PVVENSNLDTKSESAPPEKEIVVDVTDVKETE 1252
>TC227839 similar to UP|VIL4_ARATH (O65570) Villin 4, partial (12%)
Length = 673
Score = 199 bits (505), Expect(2) = 2e-56
Identities = 107/148 (72%), Positives = 116/148 (78%), Gaps = 3/148 (2%)
Frame = +3
Query: 430 KLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTS---KSNPETNSDNE 486
K YPKS D A LA KSSAI+HLTS+FE SARE LIPRS K +S KSNPET SD E
Sbjct: 57 KTYPKSMAQDTAKLATKSSAIAHLTSSFELTSARENLIPRSQKASSVTPKSNPET-SDEE 233
Query: 487 NSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEF 546
S SR ESLTIQED EGE ED+EGLPVYP+E V T STDP+ DIDVTKREAYLS EF
Sbjct: 234 GSLSSRIESLTIQEDAKEGEAEDDEGLPVYPHERVNTASTDPVEDIDVTKREAYLSSAEF 413
Query: 547 QERLGMTRSEFYKLPKWKQNKLKMAVQL 574
QE+ GM ++EFYKLPKWKQNKLKMAVQL
Sbjct: 414 QEKFGMAKNEFYKLPKWKQNKLKMAVQL 497
Score = 39.3 bits (90), Expect(2) = 2e-56
Identities = 17/19 (89%), Positives = 18/19 (94%)
Frame = +1
Query: 413 FENSNVRNLSTPPPMIRKL 431
FE+SN RNLSTPPPMIRKL
Sbjct: 7 FESSNARNLSTPPPMIRKL 63
>TC227706 similar to UP|VIL1_ARATH (O81643) Villin 1, partial (11%)
Length = 515
Score = 152 bits (384), Expect = 4e-37
Identities = 72/154 (46%), Positives = 108/154 (69%), Gaps = 6/154 (3%)
Frame = +1
Query: 135 ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQ 194
ETY E+ VALFR+QG+ P++MQAIQV+ ++SLNSSYCYILQS++ ++TW G+L+++ D
Sbjct: 1 ETYNENLVALFRVQGTSPDNMQAIQVDQVSTSLNSSYCYILQSKASIYTWIGSLSSARDH 180
Query: 195 ELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFS- 253
L +RM++L P +EG E + FW+ L K EY K ++ +DPHLF+ +
Sbjct: 181 NLLDRMVELSNPTWLPVSVREGNEPDIFWDALSGKAEYPKGKEIQGFIDDPHLFALKITR 360
Query: 254 -----EEIHNFSQDDLMTEDIFILDCHSQIFVWV 282
+EI+N++QDDL+TED+ +LDC +I+VWV
Sbjct: 361 GDFKVKEIYNYTQDDLITEDVLLLDCQREIYVWV 462
>BG507436 similar to GP|8777365|dbj| villin {Arabidopsis thaliana}, partial
(7%)
Length = 426
Score = 115 bits (289), Expect = 4e-26
Identities = 57/77 (74%), Positives = 63/77 (81%)
Frame = +2
Query: 246 HLFSCNFSEEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQD 305
+L C +EIHNFSQDDLMTEDI+ LDCHS+IFVWVGQQVD K R+QAL IGEKFLE D
Sbjct: 194 NLKQCLQVKEIHNFSQDDLMTEDIYTLDCHSEIFVWVGQQVDSKSRMQALTIGEKFLEHD 373
Query: 306 FLLETISCSAPIYIVME 322
FLLE +S APIYIV E
Sbjct: 374 FLLEGLSREAPIYIVKE 424
>TC208291 similar to UP|Q7Y0V3 (Q7Y0V3) Actin filament bundling protein
P-115-ABP, partial (8%)
Length = 723
Score = 114 bits (285), Expect = 1e-25
Identities = 54/77 (70%), Positives = 62/77 (80%)
Frame = +1
Query: 498 IQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEF 557
IQEDV E E ED E L +YPYE +K STDP+P+IDVTKRE YLS EF+E+ GM++ F
Sbjct: 1 IQEDVKEDEVEDEEXLVIYPYERLKIMSTDPVPNIDVTKRETYLSSAEFKEKFGMSKDAF 180
Query: 558 YKLPKWKQNKLKMAVQL 574
YKLPKWKQNKLKMAVQL
Sbjct: 181 YKLPKWKQNKLKMAVQL 231
>BQ629899 weakly similar to PIR|T50669|T506 villin 2 [imported] - Arabidopsis
thaliana, partial (6%)
Length = 432
Score = 70.5 bits (171), Expect = 2e-12
Identities = 42/107 (39%), Positives = 60/107 (55%), Gaps = 3/107 (2%)
Frame = +3
Query: 471 LKDTSKSNPE--TNSDNENSTGSREESLTIQEDVNEGEPE-DNEGLPVYPYESVKTDSTD 527
+K+T + PE TN D+E QE+V +G + +N V+ YE +KT S
Sbjct: 21 VKETEEVAPEAGTNGDSEQPK---------QENVEDGRNDSENNNQNVFSYEQLKTKSGS 173
Query: 528 PMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+ ID+ +REAYLS +EF+ GM + F KLP+WKQ+ LK V L
Sbjct: 174 VVSGIDLKQREAYLSDKEFETVFGMAKEAFSKLPRWKQDMLKRKVDL 314
>TC219670 weakly similar to UP|VIL2_ARATH (O81644) Villin 2, partial (6%)
Length = 678
Score = 65.5 bits (158), Expect = 6e-11
Identities = 32/70 (45%), Positives = 42/70 (59%)
Frame = +2
Query: 505 GEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWK 564
G +N V+ YE +KT S + ID+ +REAYLS +EFQ GM + F KLP+WK
Sbjct: 44 GNDSENNNQNVFSYEQLKTKSGSVVSGIDLKQREAYLSDKEFQTVFGMAKEAFSKLPRWK 223
Query: 565 QNKLKMAVQL 574
Q+ LK V L
Sbjct: 224 QDMLKRKVDL 253
>TC227707 similar to UP|VIL1_ARATH (O81643) Villin 1, partial (5%)
Length = 457
Score = 62.8 bits (151), Expect = 4e-10
Identities = 42/149 (28%), Positives = 64/149 (42%), Gaps = 51/149 (34%)
Frame = +1
Query: 198 ERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSC------- 250
+RM++L P +EG E + FW+ L K EY K ++ +DPHLF+
Sbjct: 4 DRMVELSNPTWLPVSVREGNEPDIFWDALSGKAEYPKGKEIQGFIDDPHLFALKITRGKK 183
Query: 251 --------------------------------------NFS------EEIHNFSQDDLMT 266
NF+ +EI+N++QDDL+T
Sbjct: 184 STLLRKVI*ISLGLELSWVK*QGKHV*LMVLFLTLICWNFNSRDFKVKEIYNYTQDDLIT 363
Query: 267 EDIFILDCHSQIFVWVGQQVDPKRRVQAL 295
ED+ +LDC +I+VWVG K + +AL
Sbjct: 364 EDVLLLDCQREIYVWVGLHSAVKSKQEAL 450
>AW278782 similar to PIR|T50669|T506 villin 2 [imported] - Arabidopsis
thaliana, partial (9%)
Length = 482
Score = 57.4 bits (137), Expect = 2e-08
Identities = 44/159 (27%), Positives = 73/159 (45%), Gaps = 4/159 (2%)
Frame = +3
Query: 400 RGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEP 459
R R+ A AL + F +S+ TP P R + +A L+ A + P
Sbjct: 15 RQRAEALAALNSAFNSSSGTKTFTPRPSGRG-QGSQRAAAVAALSQVLMAEKKKSPDGSP 191
Query: 460 PSAREKLIPRSLKDTSK----SNPETNSDNENSTGSREESLTIQEDVNEGEPEDNEGLPV 515
++R + + D+S+ + + + TGS + QE+ EG N+G +
Sbjct: 192 VASRSPITEETKSDSSEVEEVAEAKETEELPPETGSNGDLELKQENAEEG----NDGQRM 359
Query: 516 YPYESVKTDSTDPMPDIDVTKREAYLSPEEFQERLGMTR 554
+ YE +KT S +P +D+ +REAYLS +EF GM +
Sbjct: 360 FSYEQLKTKSGHNVPGVDLKRREAYLSEDEFNTVFGMAK 476
>TC215396 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kD chloroplast nucleoid DNA
binding protein (CND41), partial (69%)
Length = 1808
Score = 37.7 bits (86), Expect = 0.014
Identities = 32/99 (32%), Positives = 43/99 (43%)
Frame = +1
Query: 381 PEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDL 440
P S S S++ R + PA +A AAT S+ + PPP I K P + P
Sbjct: 766 PTSSTTSSSVAAKTTRASLVA-PPASSASAATPSPSS----NKPPPYIVKSSPTASPPPP 930
Query: 441 ATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNP 479
A A SA L ++ PPS P S TS ++P
Sbjct: 931 APPAASPSAPPPLPTSSTPPSPPSPAAPPSTASTSPASP 1047
>TC208290
Length = 603
Score = 34.7 bits (78), Expect = 0.12
Identities = 15/16 (93%), Positives = 15/16 (93%)
Frame = +2
Query: 559 KLPKWKQNKLKMAVQL 574
KLPKWKQNKL MAVQL
Sbjct: 2 KLPKWKQNKLXMAVQL 49
>AW598284
Length = 440
Score = 32.0 bits (71), Expect = 0.77
Identities = 17/52 (32%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Frame = +2
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREE-HLIGTWIGKNS 91
++WR+ + L D KFY GD YI + G+ + I WIGK++
Sbjct: 2 EIWRIEDFQPVPLPRPDYGKFYMGDSYIILQTTQGKGSAYLYDIHFWIGKDT 157
>TC229729 similar to GB|AAO63276.1|28950705|BT005212 At2g42260 {Arabidopsis
thaliana;} , partial (12%)
Length = 492
Score = 32.0 bits (71), Expect = 0.77
Identities = 24/77 (31%), Positives = 39/77 (50%)
Frame = +1
Query: 359 TPPLVKPKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNV 418
+PPL P+R A++ G SG S S++ S R R RGR P+ N L A + + +
Sbjct: 208 SPPLRTPQRTAAIGRGRASG-----SPGSQNTPPSTAR-RGRGRVPSRNVLPAWYPRTPL 369
Query: 419 RNLSTPPPMIRKLYPKS 435
R+++ I + +S
Sbjct: 370 RDITVVVQAIERRRARS 420
>BU548249
Length = 537
Score = 31.2 bits (69), Expect = 1.3
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = -2
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREE-HLIGTWIGKNS 91
+++WR+ + L ++ KFY GD YI + G+ + + WIGK++
Sbjct: 506 TEIWRIENFQPVPLPKSEYGKFYMGDSYIILQTTQGKGSTYFYDLHFWIGKHT 348
>TC205462 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;} , partial (54%)
Length = 1403
Score = 30.8 bits (68), Expect = 1.7
Identities = 29/120 (24%), Positives = 48/120 (39%)
Frame = +3
Query: 367 RRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPP 426
R AS+ Y GR G + + SRS S S D RSP+ + +++ + +P
Sbjct: 582 RSASLDYKGRGRGRYDDERNSRSRSRSVDSGSPARRSPSPRRSPSPQRSTSPQRSPSPRK 761
Query: 427 MIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSDNE 486
R P +++ D + P+S + PR D S+S NS+ +
Sbjct: 762 SPRGESPANRSRDGRSPTPRS------------------VSPRGRPDASRSPSPRNSNGD 887
>TC218046 weakly similar to PIR|T49196|T49196 nematode resistance-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(77%)
Length = 1617
Score = 30.8 bits (68), Expect = 1.7
Identities = 25/99 (25%), Positives = 42/99 (42%), Gaps = 11/99 (11%)
Frame = -1
Query: 396 RVRVRGRSPAFNALAATFENSNVRNL---STPPPMIRKLYPKSKTPDLATLAPKSSAISH 452
R R+ P A AT E S R+ ++ PP+ + P P + AP +S +
Sbjct: 1221 RRRISSSPPQSPASPATSEPSRTRSSKRRASSPPLESSVLPSDPPPPPISPAPCASDLCT 1042
Query: 453 LTSTFEPPSA--------REKLIPRSLKDTSKSNPETNS 483
+S PS R ++ PR+ S++ P+ +S
Sbjct: 1041 SSSVQPTPST*SRTKPFRRRRISPRADSSVSRNRPDPSS 925
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,634,978
Number of Sequences: 63676
Number of extensions: 359719
Number of successful extensions: 2228
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 2187
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2211
length of query: 574
length of database: 12,639,632
effective HSP length: 102
effective length of query: 472
effective length of database: 6,144,680
effective search space: 2900288960
effective search space used: 2900288960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC125475.5


