

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
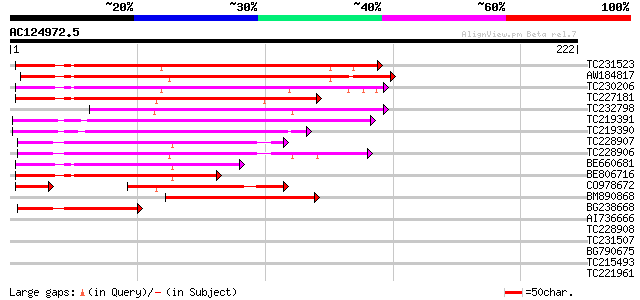
Query= AC124972.5 - phase: 0
(222 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231523 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 113 6e-26
AW184817 112 2e-25
TC230206 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 104 4e-23
TC227181 similar to UP|Q940N7 (Q940N7) AT4g05150/C17L7_70, parti... 92 2e-19
TC232798 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 91 5e-19
TC219391 similar to GB|AAO39954.1|28372944|BT003726 At5g63130 {A... 82 2e-16
TC219390 similar to GB|AAT41790.1|48310282|BT014807 At3g48240 {A... 80 7e-16
TC228907 similar to GB|AAO39954.1|28372944|BT003726 At5g63130 {A... 69 2e-12
TC228906 similar to UP|Q6ID88 (Q6ID88) At3g26510, partial (48%) 69 2e-12
BE660681 64 4e-11
BE806716 62 2e-10
CO978672 51 3e-08
BM890868 54 4e-08
BG238668 43 1e-04
AI736666 similar to GP|20197587|gb| expressed protein {Arabidops... 37 0.005
TC228908 similar to UP|Q6ID88 (Q6ID88) At3g26510, partial (21%) 37 0.007
TC231507 UP|Q8S3J3 (Q8S3J3) Hydroxyisourate hydrolase, partial (... 33 0.10
BG790675 32 0.23
TC215493 weakly similar to UP|DX_DROME (Q23985) Deltex protein, ... 32 0.23
TC221961 homologue to PIR|T00933|T00933 RNA-binding protein homo... 30 0.86
>TC231523 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (32%)
Length = 878
Score = 113 bits (283), Expect = 6e-26
Identities = 67/150 (44%), Positives = 94/150 (62%), Gaps = 6/150 (4%)
Frame = +1
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFS---D 59
+ K M +YGGKIQPR HD+ + SY+GGD KI+ VDR++ F ++KL+ S D
Sbjct: 130 KAKFMCSYGGKIQPRT---HDN-QLSYVGGDTKILAVDRSVKFPAFLSKLAAVCDSAPQD 297
Query: 60 VCFKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASF 119
+ FKYQL GED DALI + N++DL H+M EYDR+ + KP +R+FLF + +N +SF
Sbjct: 298 LTFKYQLPGEDLDALISVTNDDDLEHMMHEYDRLYRPNLKPVRMRLFLFTLSNSNPNSSF 477
Query: 120 DSLAS--VKSLNFVRV-SPFEEYSSPPTTP 146
S V++LN V S + +PP TP
Sbjct: 478 SSERDRFVEALNSGPVPSQPDPIKTPPVTP 567
>AW184817
Length = 455
Score = 112 bits (279), Expect = 2e-25
Identities = 69/155 (44%), Positives = 94/155 (60%), Gaps = 8/155 (5%)
Frame = +3
Query: 5 KLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVC--- 61
KLM ++GG IQPR HD+ +Y+ GD KI+ VDR++ F L+AKLS+ +
Sbjct: 3 KLMCSFGGSIQPRP---HDN-HLTYVSGDTKILAVDRHVKFPSLIAKLSSLANNTPSNLS 170
Query: 62 -FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASFD 120
FKYQL GED DALI + N++DL+ +M EYDR+ S +PA LR+FLFP+ N N A +
Sbjct: 171 FFKYQLPGEDLDALISVTNDDDLHQMMIEYDRLSRASPRPARLRLFLFPLHNNCNFAPTE 350
Query: 121 SLAS----VKSLNFVRVSPFEEYSSPPTTPPPPFM 151
S + V +LN V V P + PP T P F+
Sbjct: 351 SKSERQWFVDALNSVHV-PEDSPPPPPATANPDFL 452
>TC230206 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (28%)
Length = 1305
Score = 104 bits (259), Expect = 4e-23
Identities = 65/164 (39%), Positives = 98/164 (59%), Gaps = 18/164 (10%)
Frame = +3
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFS---- 58
+ K M +YGGKI PR HD+ + SY+ G+ KI+ VDR+I FS ++AKLS +
Sbjct: 171 KAKFMCSYGGKILPRS---HDN-QLSYVAGETKILAVDRSIKFSAMLAKLSALCDAPDNN 338
Query: 59 DVCFKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLF-----PVPIN 113
++ FKYQL GED DALI + N++DL+H+M EYDR+ S +P+ +R+FLF P P
Sbjct: 339 NLTFKYQLPGEDLDALISVTNDDDLDHMMHEYDRLYRASARPSRMRLFLFPSDTLPSPPP 518
Query: 114 NNKASFDSLASVKSLNFV-----RVSPFE-EYSSP---PTTPPP 148
A DS+ +++F+ + P + +++ P P PPP
Sbjct: 519 AASAPPDSVIKPNNVDFLFGLQPQPPPIDVKFNDPLPEPVAPPP 650
>TC227181 similar to UP|Q940N7 (Q940N7) AT4g05150/C17L7_70, partial (38%)
Length = 1611
Score = 91.7 bits (226), Expect = 2e-19
Identities = 53/122 (43%), Positives = 78/122 (63%), Gaps = 2/122 (1%)
Frame = +3
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFM-FSDVC 61
R++ M ++GGKI PR HD+ + Y+GGD +I+ V+R+I FS L+ KLS S++
Sbjct: 159 RIRFMCSFGGKILPRP---HDN-QLRYVGGDTRIVAVNRSITFSALILKLSKLSGMSNIT 326
Query: 62 FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSR-KPAWLRVFLFPVPINNNKASFD 120
KYQL ED DALI + +ED+ ++M EYDR+ H + A LR+FLFP ++ +S
Sbjct: 327 AKYQLPNEDLDALISVTTDEDVENMMDEYDRVAHNQNPRSARLRLFLFPEGEDSRASSIS 506
Query: 121 SL 122
SL
Sbjct: 507 SL 512
>TC232798 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (10%)
Length = 707
Score = 90.5 bits (223), Expect = 5e-19
Identities = 51/122 (41%), Positives = 73/122 (59%), Gaps = 5/122 (4%)
Frame = +1
Query: 32 GDNKIITVDRNINFSDLMAKLSTF---MFSDVCFKYQLLGEDFDALIPIYNEEDLNHIMF 88
GD KI+ VDR+I F ++AKLS +++ FKYQL GED DALI + N++DL+H+M
Sbjct: 1 GDTKILAVDRSIKFPAMLAKLSALCDAQDNNITFKYQLPGEDLDALISVTNDDDLDHMMH 180
Query: 89 EYDRMCHFSRKPAWLRVFLFP--VPINNNKASFDSLASVKSLNFVRVSPFEEYSSPPTTP 146
EYDR+ S +P+ +R+FLFP P + AS +K N + E+ + P P
Sbjct: 181 EYDRLYRASARPSRMRLFLFPSDTPPSPPPASAPPDTVIKPNNVDFLFALEKNVAVPAPP 360
Query: 147 PP 148
PP
Sbjct: 361 PP 366
>TC219391 similar to GB|AAO39954.1|28372944|BT003726 At5g63130 {Arabidopsis
thaliana;} , partial (44%)
Length = 718
Score = 82.0 bits (201), Expect = 2e-16
Identities = 55/142 (38%), Positives = 75/142 (52%)
Frame = +1
Query: 2 ERVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVC 61
+ +KL+ +YGGKI PR T LRY+ GG +++TV R+I+FS+LM KLS F S V
Sbjct: 142 QTIKLLCSYGGKILPRAT--DGELRYA--GGHTRVLTVARSISFSELMVKLSEFCGSSVI 309
Query: 62 FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASFDS 121
+ QL D + LI I N+EDL I+ EYDR P ++ L P + S S
Sbjct: 310 LRCQLPKGDLETLISITNDEDLASIIEEYDRASLKLAHPLKIKAVLSPPKSSKKTPSAPS 489
Query: 122 LASVKSLNFVRVSPFEEYSSPP 143
AS + + SP S P
Sbjct: 490 SASSSASHSPSRSPHTSTDSLP 555
>TC219390 similar to GB|AAT41790.1|48310282|BT014807 At3g48240 {Arabidopsis
thaliana;} , partial (53%)
Length = 845
Score = 80.1 bits (196), Expect = 7e-16
Identities = 50/117 (42%), Positives = 68/117 (57%)
Frame = +1
Query: 2 ERVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVC 61
+ +KL+ +YGGKI PR T LRY +GG +++TVDR+I+F +LM KL F S V
Sbjct: 148 QTIKLLCSYGGKILPRAT--DGELRY--VGGHTRVLTVDRSISFPELMVKLRVFCGSSVI 315
Query: 62 FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKAS 118
+ QL D + LI I N+EDL I+ EYDR P ++ L P P + KAS
Sbjct: 316 LRCQLPKGDLETLISITNDEDLASIIEEYDRSSLKLAHPLKIKAVLSP-PKSMKKAS 483
>TC228907 similar to GB|AAO39954.1|28372944|BT003726 At5g63130 {Arabidopsis
thaliana;} , partial (53%)
Length = 537
Score = 68.6 bits (166), Expect = 2e-12
Identities = 40/107 (37%), Positives = 63/107 (58%), Gaps = 1/107 (0%)
Frame = +1
Query: 4 VKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVCF- 62
+K + +YGGKI PR + + Y+GG +++ VDR+I FS+L+ KL + V +
Sbjct: 178 LKFLCSYGGKILPR----YPDGKLRYLGGHTRVLAVDRSIPFSELLLKLEELCGASVRYL 345
Query: 63 KYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFP 109
+ QL ED DAL+ I ++EDL +++ EYDR+ +R FL P
Sbjct: 346 RCQLPSEDLDALVSITSDEDLANLIEEYDRVSSLK-----IRAFLSP 471
>TC228906 similar to UP|Q6ID88 (Q6ID88) At3g26510, partial (48%)
Length = 927
Score = 68.6 bits (166), Expect = 2e-12
Identities = 52/150 (34%), Positives = 76/150 (50%), Gaps = 11/150 (7%)
Frame = +2
Query: 4 VKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVC-F 62
+K + +YGGKI PR + + Y+GG +I+ VDR+I FS+L+ KL + V
Sbjct: 155 LKFLCSYGGKILPR----YPDGKLRYLGGHTRILAVDRSIPFSELLLKLEELCGASVRHL 322
Query: 63 KYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFP--------VPINN 114
+ QL ED DAL+ I ++EDL +++ EYDR+ +R FL P P +
Sbjct: 323 RCQLPSEDLDALVSITSDEDLANLIEEYDRVSSLK-----IRAFLSPPTPPTPPTPPRSP 487
Query: 115 NKASF--DSLASVKSLNFVRVSPFEEYSSP 142
NK S AS+ S + S Y SP
Sbjct: 488 NKFSTAPSKSASISSSSASSSSSSSSYYSP 577
>BE660681
Length = 541
Score = 64.3 bits (155), Expect = 4e-11
Identities = 38/92 (41%), Positives = 55/92 (59%), Gaps = 2/92 (2%)
Frame = +3
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVCF 62
+++LM +YGG I PR HD Y+GGD +II +R + +DL +LS + F
Sbjct: 129 KLRLMCSYGGHIVPRP---HDK-SLCYVGGDTRIIVSERATSLADLSMRLSKTFLNGRPF 296
Query: 63 --KYQLLGEDFDALIPIYNEEDLNHIMFEYDR 92
KYQL ED D+LI + +EDL +++ EYDR
Sbjct: 297 TLKYQLPNEDLDSLISVTTDEDLENMIDEYDR 392
>BE806716
Length = 407
Score = 62.0 bits (149), Expect = 2e-10
Identities = 36/83 (43%), Positives = 52/83 (62%), Gaps = 2/83 (2%)
Frame = +1
Query: 3 RVKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVCF 62
+++LM +YGG I PR HD SYIGGD +I+ VDR+ + DL ++LS + + F
Sbjct: 169 KLRLMCSYGGHIMPRP---HDK-SLSYIGGDTRIVVVDRHSSLKDLCSRLSRTILNGRPF 336
Query: 63 --KYQLLGEDFDALIPIYNEEDL 83
KYQL ED ++LI + +EDL
Sbjct: 337 TLKYQLPNEDLESLITVTTDEDL 405
>CO978672
Length = 725
Score = 50.8 bits (120), Expect(2) = 3e-08
Identities = 28/65 (43%), Positives = 40/65 (61%), Gaps = 2/65 (3%)
Frame = -3
Query: 47 DLMAKLSTFM--FSDVCFKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLR 104
+LM K+S+ + D+ KYQL+ ED DAL+ + EED+ H++ E+DR A LR
Sbjct: 621 ELMKKVSSMVEGVGDMVLKYQLIPEDLDALVSVRTEEDVKHMIEEHDR----HHTGALLR 454
Query: 105 VFLFP 109
FLFP
Sbjct: 453 AFLFP 439
Score = 23.9 bits (50), Expect(2) = 3e-08
Identities = 8/15 (53%), Positives = 12/15 (79%)
Frame = -2
Query: 3 RVKLMITYGGKIQPR 17
+VK + +YGGK+ PR
Sbjct: 685 KVKFLCSYGGKVLPR 641
>BM890868
Length = 421
Score = 54.3 bits (129), Expect = 4e-08
Identities = 27/60 (45%), Positives = 39/60 (65%)
Frame = +1
Query: 62 FKYQLLGEDFDALIPIYNEEDLNHIMFEYDRMCHFSRKPAWLRVFLFPVPINNNKASFDS 121
F +L GED DALI + N++DL H+M EY+R+ + KP +R+FLF + N +SF S
Sbjct: 16 FLSKLAGEDLDALISVTNDDDLEHMMLEYERLYRPNLKPIRMRLFLFTLSN*NPNSSFYS 195
>BG238668
Length = 442
Score = 42.7 bits (99), Expect = 1e-04
Identities = 21/49 (42%), Positives = 32/49 (64%)
Frame = +2
Query: 4 VKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKL 52
+K + +YGGKI PR + + Y+GG +I+ VDR+I FS+L+ KL
Sbjct: 170 LKFLCSYGGKILPR----YPDGKLRYLGGHTRILAVDRSIPFSELLLKL 304
>AI736666 similar to GP|20197587|gb| expressed protein {Arabidopsis
thaliana}, partial (6%)
Length = 357
Score = 37.4 bits (85), Expect = 0.005
Identities = 27/90 (30%), Positives = 44/90 (48%), Gaps = 10/90 (11%)
Frame = +3
Query: 29 YIGGDNKIITVDRNINFSDLMAKLSTFMFSDVC-------FKYQLLGEDFDALIPIYNEE 81
Y+GGD +II +R + DL ST +++ ED + LI + +E
Sbjct: 18 YVGGDTRIIGCERATSLWDL----STAPINNIA*MEDP*RSSTSCATEDLECLIYVTTDE 185
Query: 82 DLNHIMFEYDRMCHFSR---KPAWLRVFLF 108
DL +++ EY+R + KP +R+FLF
Sbjct: 186 DLENMIEEYERTACVATCCDKPCRIRLFLF 275
>TC228908 similar to UP|Q6ID88 (Q6ID88) At3g26510, partial (21%)
Length = 775
Score = 37.0 bits (84), Expect = 0.007
Identities = 19/61 (31%), Positives = 33/61 (53%)
Frame = +3
Query: 4 VKLMITYGGKIQPRLTALHDHLRYSYIGGDNKIITVDRNINFSDLMAKLSTFMFSDVCFK 63
+K + +YGGKI PR + + Y+GG +++ VDR+I FS + T ++ F
Sbjct: 516 LKFLCSYGGKILPR----YPDGKLRYLGGHTRVLAVDRSIPFSGQFSLSHTHSLTN*IFN 683
Query: 64 Y 64
+
Sbjct: 684 F 686
>TC231507 UP|Q8S3J3 (Q8S3J3) Hydroxyisourate hydrolase, partial (87%)
Length = 1461
Score = 33.1 bits (74), Expect = 0.10
Identities = 18/75 (24%), Positives = 33/75 (44%)
Frame = +1
Query: 124 SVKSLNFVRVSPFEEYSSPPTTPPPPFMATKDTVTKAEMKKFRGLHVVNDEFEDILKLYL 183
+V N + +++ +SPP PPF AT DT+ + + + +H + +LY
Sbjct: 589 TVNEPNVFALGGYDQGNSPPRRCSPPFCATNDTMGNSTYEPYLAVHHILLSHSSAARLYW 768
Query: 184 TCNFSKVKDFHGLHV 198
K F G+ +
Sbjct: 769 RKYRDKQHGFVGISI 813
>BG790675
Length = 417
Score = 32.0 bits (71), Expect = 0.23
Identities = 16/30 (53%), Positives = 19/30 (63%), Gaps = 2/30 (6%)
Frame = +1
Query: 136 FEEYSSPPTTPPPPFM--ATKDTVTKAEMK 163
F+E S PP TPPP + ATK TKA+ K
Sbjct: 253 FKENSLPPLTPPPSYFQNATKKATTKAKNK 342
>TC215493 weakly similar to UP|DX_DROME (Q23985) Deltex protein, partial (6%)
Length = 2072
Score = 32.0 bits (71), Expect = 0.23
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 3/40 (7%)
Frame = +3
Query: 73 ALIPIYNEEDLNHIMFEYDRMCHFSR---KPAWLRVFLFP 109
+LI + +EDL +++ EYDR + KP+ +R+FLFP
Sbjct: 3 SLISVTTDEDLENMIDEYDRTAAAATSAVKPSRIRLFLFP 122
>TC221961 homologue to PIR|T00933|T00933 RNA-binding protein homolog
At2g42240 - Arabidopsis thaliana {Arabidopsis thaliana;}
, partial (29%)
Length = 450
Score = 30.0 bits (66), Expect = 0.86
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = +2
Query: 135 PFEEYSSPPTTPPPPFMA 152
P++ Y PP PPPP MA
Sbjct: 80 PYQYYQQPPPPPPPPMMA 133
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,308,593
Number of Sequences: 63676
Number of extensions: 177338
Number of successful extensions: 3544
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 2465
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3363
length of query: 222
length of database: 12,639,632
effective HSP length: 94
effective length of query: 128
effective length of database: 6,654,088
effective search space: 851723264
effective search space used: 851723264
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC124972.5


