

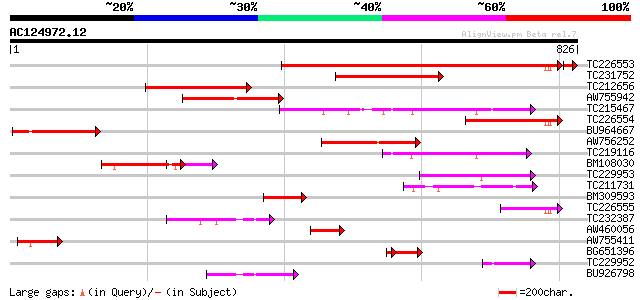
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124972.12 - phase: 0 /pseudo
(826 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC226553 similar to UP|Q9SDG8 (Q9SDG8) ESTs AU068544(C30430), pa... 602 e-172
TC231752 similar to UP|Q9SDG8 (Q9SDG8) ESTs AU068544(C30430), pa... 254 1e-67
TC212656 similar to GB|AAC77862.2|20197419|AC005623 Argonaute (A... 252 4e-67
AW755942 similar to GP|20197419|gb Argonaute (AGO1)-like protein... 245 7e-65
TC215467 similar to UP|AGO1_ARATH (O04379) Argonaute protein, pa... 219 3e-57
TC226554 similar to UP|Q9SDG8 (Q9SDG8) ESTs AU068544(C30430), pa... 192 5e-49
BU964667 weakly similar to GP|20197419|gb| Argonaute (AGO1)-like... 182 5e-46
AW756252 weakly similar to PIR|T01113|T01 translation initiation... 170 3e-42
TC219116 homologue to UP|PINH_ARATH (Q9XGW1) PINHEAD protein (ZW... 165 7e-41
BM108030 151 1e-36
TC229953 similar to UP|Q8LP00 (Q8LP00) ZLL/PNH homologous protei... 133 4e-31
TC211731 similar to UP|Q9SHF2 (Q9SHF2) T19E23.8, partial (13%) 125 1e-28
BM309593 111 1e-24
TC226555 similar to GB|AAC77862.2|20197419|AC005623 Argonaute (A... 102 7e-22
TC232387 similar to UP|AGO1_ARATH (O04379) Argonaute protein, pa... 91 2e-18
AW460056 83 4e-16
AW755411 weakly similar to GP|20197419|gb| Argonaute (AGO1)-like... 79 1e-14
BG651396 similar to PIR|T01113|T011 translation initiation facto... 69 3e-14
TC229952 homologue to UP|Q8LP00 (Q8LP00) ZLL/PNH homologous prot... 64 3e-10
BU926798 homologue to SP|Q9XGW1|PINH PINHEAD protein (ZWILLE pro... 63 5e-10
>TC226553 similar to UP|Q9SDG8 (Q9SDG8) ESTs AU068544(C30430), partial (54%)
Length = 1899
Score = 602 bits (1552), Expect(2) = e-172
Identities = 310/444 (69%), Positives = 351/444 (78%), Gaps = 35/444 (7%)
Frame = +3
Query: 397 LVEKSRQKPVERMRVLSNALKASNYGSEPMLRNCGISITSEFTQVDGRVLQAPRLKFGN- 455
LVEKSRQKP ERM++LS+AL+ SNYG+EPMLRNCGISI++ FT+V+GRVL APRLKFGN
Sbjct: 3 LVEKSRQKPQERMKILSDALRTSNYGAEPMLRNCGISISTGFTEVEGRVLPAPRLKFGNG 182
Query: 456 EDFNPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCDVRGLVRDLIKCGGMKGILVEQPKD 515
ED NPRNGRWN + KFVEP + W+V NFSARCDVRGLVRDLI+ G MKGI +EQP D
Sbjct: 183 EDLNPRNGRWNVSRVKFVEPSKIERWAVANFSARCDVRGLVRDLIRIGDMKGITIEQPFD 362
Query: 516 VIEENRQFKGEPPVFRVEKMFADVL-KLSKRPSFLLCLLPERKNSDLYGPWKKKNLAEFG 574
V +EN QF+ PP+ RVEKMF + KL P FLLCLLP+RKN D+YGPWKKKNLA+FG
Sbjct: 363 VFDENPQFRRAPPMVRVEKMFEHIQSKLPGAPQFLLCLLPDRKNCDIYGPWKKKNLADFG 542
Query: 575 IVTQCIAPTRVNDQYLTNVLLKINAKLGGMNSWLGVEHSRSIPIVSKVPTLILGMDVSHG 634
I+ QC+ P RVNDQYLTNV+LKINAKLGG+NS LGVEHS S+P+VSK PTLILGMDVSHG
Sbjct: 543 IINQCMCPLRVNDQYLTNVMLKINAKLGGLNSLLGVEHSPSLPVVSKAPTLILGMDVSHG 722
Query: 635 SPGQPDIPSIAAVVSSRKWPLISKYRACVRTQGSKVEMIDNLFKPVSDKEDEGIIRELLL 694
SPGQ DIPSIAAVVSSR WPLISKYRACVRTQ +K+EMIDNLFK VS+KEDEGIIRELLL
Sbjct: 723 SPGQTDIPSIAAVVSSRHWPLISKYRACVRTQSAKMEMIDNLFKLVSEKEDEGIIRELLL 902
Query: 695 DFFHSSEERRPENIIIFRDGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKN 754
DF+ +S R+PENIIIFRDGVSESQFN+VLN+EL +IIEACKFLDENW PKF+VIVAQKN
Sbjct: 903 DFYTTSGRRKPENIIIFRDGVSESQFNQVLNIELDRIIEACKFLDENWEPKFVVIVAQKN 1082
Query: 755 HHTKFFQPRSPDNVPPAKFAILGIM------IFIC---------------VLM------- 786
HHT+FFQP SPDNVPP I ++C VL+
Sbjct: 1083HHTRFFQPGSPDNVPPGTVIDNKICHPRNYDFYLCAHAGMIGTSRPTHYHVLLDQVGFSP 1262
Query: 787 -----LE*FLSYVYQRSTTAISVV 805
L LSYVYQRSTTAISVV
Sbjct: 1263DQLQELVHSLSYVYQRSTTAISVV 1334
Score = 21.9 bits (45), Expect(2) = e-172
Identities = 10/19 (52%), Positives = 12/19 (62%)
Frame = +1
Query: 808 YMQLLQSAMHTWLHLKLDS 826
++ LLQ AM TWL L S
Sbjct: 1324 FLLLLQYAMRTWLLLSWGS 1380
>TC231752 similar to UP|Q9SDG8 (Q9SDG8) ESTs AU068544(C30430), partial (17%)
Length = 476
Score = 254 bits (649), Expect = 1e-67
Identities = 126/158 (79%), Positives = 136/158 (85%), Gaps = 1/158 (0%)
Frame = +2
Query: 475 PVSLGNWSVVNFSARCDVRGLVRDLIKCGGMKGILVEQPKDVIEENRQFKGEPPVFRVEK 534
P + W+VVNFSARCD RGLVRDLIKCGGMKGI+++QP DV EEN QF+ PPV RVEK
Sbjct: 2 PTKIERWAVVNFSARCDTRGLVRDLIKCGGMKGIVIDQPFDVFEENGQFRRAPPVVRVEK 181
Query: 535 MFADVL-KLSKRPSFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTRVNDQYLTNV 593
MF V KL P FLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTRVNDQYLTNV
Sbjct: 182 MFELVQSKLPGAPQFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPTRVNDQYLTNV 361
Query: 594 LLKINAKLGGMNSWLGVEHSRSIPIVSKVPTLILGMDV 631
LLKINAKLGG+NS LGVEHS SIPIVS+ PT+I+GMDV
Sbjct: 362 LLKINAKLGGLNSILGVEHSPSIPIVSRAPTIIIGMDV 475
>TC212656 similar to GB|AAC77862.2|20197419|AC005623 Argonaute (AGO1)-like
protein {Arabidopsis thaliana;} , partial (14%)
Length = 465
Score = 252 bits (644), Expect = 4e-67
Identities = 122/155 (78%), Positives = 133/155 (85%)
Frame = +1
Query: 198 LDIILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCKGLHSSFRTTQSGLSLNIDVS 257
LDIILRQH+A QGCLLVRQ+FFHN+PNN DVGGGVL C+G HSSFRTTQSGLSLNIDVS
Sbjct: 1 LDIILRQHAANQGCLLVRQSFFHNNPNNFADVGGGVLGCRGFHSSFRTTQSGLSLNIDVS 180
Query: 258 TTMIVRPGPVVDFLIENQNVRDPFSLDWNKAKRTLKNLRITAKPSNQEYKITGLSELSCK 317
TTMI+ PGPVVDFLI NQNVRDPF LDW KAKRTLKNLRI PSNQE+KI+GLSEL C+
Sbjct: 181 TTMIISPGPVVDFLISNQNVRDPFQLDWAKAKRTLKNLRIKTSPSNQEFKISGLSELPCR 360
Query: 318 DQLFTMKKRGAVAGEDDTEEITVYDYFVHRRKIDL 352
+Q FT+K +G GED EEITVYDYFV RKIDL
Sbjct: 361 EQTFTLKGKGGGDGEDGNEEITVYDYFVKVRKIDL 465
>AW755942 similar to GP|20197419|gb Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (14%)
Length = 438
Score = 245 bits (625), Expect = 7e-65
Identities = 122/148 (82%), Positives = 134/148 (90%), Gaps = 1/148 (0%)
Frame = +2
Query: 253 NIDVSTTMIVRPGPVVDFLIENQNVRDPFSLDWNKAKRTLKNLRITAKPSNQEYKITGLS 312
NIDVSTTMI+ PGPVVDFLI NQNVRDPFSLDW KAKRTLKNLRI + PSNQE+KITGLS
Sbjct: 2 NIDVSTTMIITPGPVVDFLISNQNVRDPFSLDWAKAKRTLKNLRIKSSPSNQEFKITGLS 181
Query: 313 ELSCKDQLFTMKKRGAVAGEDDT-EEITVYDYFVHRRKIDLQYSAGLPCINVGKPKRPTY 371
EL CKDQ+FT+KK+G G+DDT EE+TVYDYFV+ RKIDL+YS LPCINVGKPKRPTY
Sbjct: 182 ELPCKDQMFTLKKKG---GDDDTEEEVTVYDYFVNIRKIDLRYSGDLPCINVGKPKRPTY 352
Query: 372 IPIELCSLISLQRYTKALSTSQRSSLVE 399
IP+ELCSL+SLQRYTKALST QRSSLVE
Sbjct: 353 IPLELCSLVSLQRYTKALSTLQRSSLVE 436
>TC215467 similar to UP|AGO1_ARATH (O04379) Argonaute protein, partial (51%)
Length = 1966
Score = 219 bits (559), Expect = 3e-57
Identities = 145/398 (36%), Positives = 209/398 (52%), Gaps = 24/398 (6%)
Frame = +2
Query: 393 QRSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLRNCGISITSEFTQVDGRVLQAPRLK 452
Q ++L+ + Q+P ER R + + + Y +P + GI I+ + QV+ R+L AP LK
Sbjct: 8 QITNLLRVTCQRPGERERDIMQTVHHNAYHEDPYAKEFGIKISEKLAQVEARILPAPWLK 187
Query: 453 FGN----EDFNPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCD---VRGLVRDLIKCGGM 505
+ + +D P+ G+WN NKK V ++ NW +NFS RG +L + +
Sbjct: 188 YHDTGREKDCLPQVGQWNMMNKKMVNGGTVNNWFCINFSRNVQDSVARGFCYELAQMCYI 367
Query: 506 KGILVEQPKDVIEENRQFKGEPPVFRVEKMFADVLKL----------SKRPSFLLCLLPE 555
G+ P+ V+ PPV VLK K L+ +LP+
Sbjct: 368 SGMAFT-PEPVV---------PPVSARPDQVEKVLKTRYHDAKNKLQGKELDLLIVILPD 517
Query: 556 RKNSDLYGPWKKKNLAEFGIVTQCIAPTRV---NDQYLTNVLLKINAKLGGMNSWLGVEH 612
N LYG K+ + G+V+QC V + QYL NV LKIN K+GG N+ L
Sbjct: 518 N-NGSLYGDLKRICETDLGLVSQCCLTKHVFKMSKQYLANVALKINVKVGGRNTVLVDAL 694
Query: 613 SRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQGSKVEM 672
SR IP+VS PT+I G DV+H PG+ PSIAAVV+S+ +P I+KY V Q + E+
Sbjct: 695 SRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDYPEITKYAGLVCAQAHRQEL 874
Query: 673 IDNLFK----PVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNVEL 728
I +LFK PV G+I+ELL+ F ++ + +P+ II +RDGVSE QF +VL EL
Sbjct: 875 IQDLFKQWQDPVRGTVTGGMIKELLISFRRATGQ-KPQRIIFYRDGVSEGQFYQVLLFEL 1051
Query: 729 SQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPD 766
I +AC L+ N+ P +V QK HHT+ F D
Sbjct: 1052DAIRKACASLEPNYQPPVTFVVVQKRHHTRLFASNHHD 1165
>TC226554 similar to UP|Q9SDG8 (Q9SDG8) ESTs AU068544(C30430), partial (25%)
Length = 944
Score = 192 bits (488), Expect = 5e-49
Identities = 108/174 (62%), Positives = 120/174 (68%), Gaps = 33/174 (18%)
Frame = +3
Query: 665 TQGSKVEMIDNLFKPVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVL 724
TQ K+EMIDNLFK VSDKEDEGI+RELLLDF+ SS R+P+NIIIFRDGVSESQFN+VL
Sbjct: 3 TQSPKMEMIDNLFKKVSDKEDEGIMRELLLDFYTSSGNRKPDNIIIFRDGVSESQFNQVL 182
Query: 725 NVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPDNVPPAKFAILGIM----- 779
N+EL QIIEACKFLDE WNPKF+VIVAQKNHHTKFFQP +PDNVPP I
Sbjct: 183 NIELDQIIEACKFLDEKWNPKFLVIVAQKNHHTKFFQPGAPDNVPPGTVIDNKICHPRNY 362
Query: 780 -IFIC---------------VLM------------LE*FLSYVYQRSTTAISVV 805
++C VL+ L LSYVYQRSTTAISVV
Sbjct: 363 DFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAISVV 524
>BU964667 weakly similar to GP|20197419|gb| Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (10%)
Length = 444
Score = 182 bits (462), Expect = 5e-46
Identities = 89/129 (68%), Positives = 107/129 (81%), Gaps = 1/129 (0%)
Frame = +1
Query: 5 ENKECLPPPPPIVPPNVKPIKIEQEHLKKKLPTKAPMARRGLGTKGAKLPLLTNHFEVNV 64
EN++ LPPPPP+VP +V P+K E E KK ++ P+ARRGL +KG KL LLTNH+ VNV
Sbjct: 67 ENEDSLPPPPPVVPSDVVPVKAEPE---KKKASRFPIARRGLASKGTKLQLLTNHYRVNV 237
Query: 65 ANTNRVFFQYSVALFYEDGRPVEGKGAGRKIIDKVQETYDSELNGKDLAYDGE-TLFTIG 123
ANT+ F+QYSVALFY+DGRPVEGKG GRK++D+V ETYDSELNGKD AYDGE TLFT+G
Sbjct: 238 ANTDGHFYQYSVALFYDDGRPVEGKGVGRKLLDRVHETYDSELNGKDFAYDGEKTLFTLG 417
Query: 124 SLAQKKLEF 132
SLA+ KLEF
Sbjct: 418 SLARNKLEF 444
>AW756252 weakly similar to PIR|T01113|T01 translation initiation factor
eIF-2C homolog T21L14.12 - Arabidopsis thaliana, partial
(14%)
Length = 435
Score = 170 bits (430), Expect = 3e-42
Identities = 82/146 (56%), Positives = 106/146 (72%), Gaps = 2/146 (1%)
Frame = +1
Query: 455 NEDFNPRNGRWNFNNKKFVEPVSLGNWSVVNFSARCDVRGLVRDLIKCGGMKGILVEQPK 514
N+D P NGRWNFN K ++ + W+VVNFSA CD + R+LI+CG KGI +E+P
Sbjct: 1 NDDCIPHNGRWNFNKKTLLQASHIDYWAVVNFSASCDTSYISRELIRCGMSKGINIERPY 180
Query: 515 DVIEENRQFKGEPPVFRVEKMFADVL--KLSKRPSFLLCLLPERKNSDLYGPWKKKNLAE 572
+IEE Q + PV RVE+MF D+L KL++ P +LC+LPERK D+YGPWKKK L+E
Sbjct: 181 TLIEEEPQLRKSHPVARVERMF-DLLASKLNREPKLILCVLPERKICDIYGPWKKKCLSE 357
Query: 573 FGIVTQCIAPTRVNDQYLTNVLLKIN 598
G+VTQCIAP ++ +QYLTNVLLKIN
Sbjct: 358 IGVVTQCIAPVKITNQYLTNVLLKIN 435
>TC219116 homologue to UP|PINH_ARATH (Q9XGW1) PINHEAD protein (ZWILLE
protein), partial (40%)
Length = 1551
Score = 165 bits (418), Expect = 7e-41
Identities = 95/225 (42%), Positives = 132/225 (58%), Gaps = 7/225 (3%)
Frame = +3
Query: 543 SKRPSFLLCLLPERKNSDLYGPWKKKNLAEFGIVTQCIAPT---RVNDQYLTNVLLKINA 599
+K LL +LP+ N LYG K+ + G+++QC ++ QYL NV LKIN
Sbjct: 126 AKELELLLAILPDN-NGSLYGDLKRICETDLGLISQCCLTKHVFKITKQYLANVSLKINV 302
Query: 600 KLGGMNSWLGVEHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKY 659
K+GG N+ L S IP+VS +PT+I G DV+H G+ PSIAAVV+S+ WP ++KY
Sbjct: 303 KMGGRNTVLLDAVSCRIPLVSDIPTIIFGADVTHPENGEDSSPSIAAVVASQDWPEVTKY 482
Query: 660 RACVRTQGSKVEMIDNLFK----PVSDKEDEGIIRELLLDFFHSSEERRPENIIIFRDGV 715
V Q + E+I +L+K PV G+IR+LL+ F + ++P II +RDGV
Sbjct: 483 AGLVCAQAHRQELIQDLYKTWQDPVRGTVSGGMIRDLLVS-FRKATGQKPLRIIFYRDGV 659
Query: 716 SESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFF 760
SE QF +VL EL I +AC L+ N+ P IV QK HHT+ F
Sbjct: 660 SEGQFYQVLLYELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLF 794
>BM108030
Length = 566
Score = 151 bits (381), Expect = 1e-36
Identities = 83/135 (61%), Positives = 100/135 (73%), Gaps = 12/135 (8%)
Frame = +2
Query: 134 VVVEDVASNRNNANTSPD--------KKRIRKSYRSKTYKVEINFAKEIPLQAIANALKG 185
VV+EDV SNRNN N SPD +KR+R+ YRSK++KVEI+FA +IP+QAIA+AL+G
Sbjct: 5 VVLEDVTSNRNNGNCSPDGLGDNESDRKRMRRPYRSKSFKVEISFAAKIPMQAIASALRG 184
Query: 186 HEAENYQEAIRVLDIILRQHSAKQGCLLVRQNFFHNDPNNLNDVGGGVLSCKGL----HS 241
E EN+QEAIRVLDIILRQH+AKQGCLLVRQ+FFHN+PNN DV C L
Sbjct: 185 QETENFQEAIRVLDIILRQHAAKQGCLLVRQSFFHNNPNNFADVRXW---CPRL*XIPLQ 355
Query: 242 SFRTTQSGLSLNIDV 256
+ +SGLSLNIDV
Sbjct: 356 ALELHRSGLSLNIDV 400
Score = 43.5 bits (101), Expect = 4e-04
Identities = 28/77 (36%), Positives = 38/77 (48%), Gaps = 3/77 (3%)
Frame = +3
Query: 229 VGGGVLSCKGLHSSF-RTTQSGLSLNIDVSTTMIVRPGPV--VDFLIENQNVRDPFSLDW 285
+G GVL C+ HS T+ L ST + P + + +++ PF LDW
Sbjct: 315 LGXGVLGCRXFHSKL*NYTEVACLLT*MFSTYXDISPWACXWISLISQSKXXXFPFPLDW 494
Query: 286 NKAKRTLKNLRITAKPS 302
KAKRT KNL+I PS
Sbjct: 495 AKAKRTPKNLKI*TXPS 545
>TC229953 similar to UP|Q8LP00 (Q8LP00) ZLL/PNH homologous protein, partial
(18%)
Length = 537
Score = 133 bits (334), Expect = 4e-31
Identities = 76/174 (43%), Positives = 102/174 (57%), Gaps = 4/174 (2%)
Frame = +3
Query: 597 INAKLGGMNSWLGVEHSRSIPIVSKVPTLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLI 656
IN K+GG N+ L S IP+VS +PT+I G DV+H G+ PSIAAVV+S+ WP +
Sbjct: 3 INVKMGGRNTVLLDALSWRIPLVSDIPTIIFGADVTHPESGEDPCPSIAAVVASQDWPEV 182
Query: 657 SKYRACVRTQGSKVEMIDNLFKPVSDKED----EGIIRELLLDFFHSSEERRPENIIIFR 712
+KY V Q + E+I +LFK D G+IRELLL F + ++P II +R
Sbjct: 183 TKYAGLVCAQPHREELIQDLFKCWKDPHHGIVYGGMIRELLLS-FKKATGQKPLRIIFYR 359
Query: 713 DGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPD 766
DGVSE QF +VL EL I +AC L+ ++ P +V QK HHT+ F D
Sbjct: 360 DGVSEGQFYQVLLYELDAIRKACASLEPSYQPPVTFVVVQKRHHTRLFSNNHDD 521
>TC211731 similar to UP|Q9SHF2 (Q9SHF2) T19E23.8, partial (13%)
Length = 594
Score = 125 bits (313), Expect = 1e-28
Identities = 80/205 (39%), Positives = 110/205 (53%), Gaps = 10/205 (4%)
Frame = +3
Query: 574 GIVTQCIAPTRVN---DQYLTNVLLKINAKLGGMNSWLGVEHSRSIPIVSKVP------- 623
GIVTQC N DQYLTN+ LKINAK+GG N + +++++P
Sbjct: 30 GIVTQCCLSGIANEGKDQYLTNLALKINAKIGGSN----------VELINRLPHFEGEGH 179
Query: 624 TLILGMDVSHGSPGQPDIPSIAAVVSSRKWPLISKYRACVRTQGSKVEMIDNLFKPVSDK 683
+ +G DV+H + + PSIAAVV++ WP ++Y A V QG +VE I N
Sbjct: 180 VMFIGADVNHPASRDINSPSIAAVVATVNWPAANRYAARVCAQGHRVEKILNF------- 338
Query: 684 EDEGIIRELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNVELSQIIEACKFLDENWN 743
G I L+ ++ + RPE I++FRDGVSESQF+ VL EL + F D N+
Sbjct: 339 ---GRICYELVSYYDRLNKVRPEKIVVFRDGVSESQFHMVLTEELQDLKSV--FSDANYF 503
Query: 744 PKFMVIVAQKNHHTKFFQPRSPDNV 768
P +IVAQK H T+FF D +
Sbjct: 504 PTITIIVAQKRHQTRFFPVGPKDGI 578
>BM309593
Length = 188
Score = 111 bits (278), Expect = 1e-24
Identities = 54/62 (87%), Positives = 60/62 (96%)
Frame = +2
Query: 371 YIPIELCSLISLQRYTKALSTSQRSSLVEKSRQKPVERMRVLSNALKASNYGSEPMLRNC 430
YIP+ELCSL+SLQRYTKALST QR+SLVEKSRQKP ERMRVL++ALK+SNYGSEPMLRNC
Sbjct: 2 YIPLELCSLVSLQRYTKALSTLQRASLVEKSRQKPQERMRVLTDALKSSNYGSEPMLRNC 181
Query: 431 GI 432
GI
Sbjct: 182 GI 187
>TC226555 similar to GB|AAC77862.2|20197419|AC005623 Argonaute (AGO1)-like
protein {Arabidopsis thaliana;} , partial (17%)
Length = 899
Score = 102 bits (254), Expect = 7e-22
Identities = 64/123 (52%), Positives = 71/123 (57%), Gaps = 33/123 (26%)
Frame = +1
Query: 716 SESQFNEVLNVELSQIIEACKFLDENWNPKFMVIVAQKNHHTKFFQPRSPDNVPPAKFAI 775
SESQFN+VLN+ QIIEACKFLDE PKF+VIVAQKNHHTKFFQP +PDNVPP
Sbjct: 1 SESQFNQVLNIXXDQIIEACKFLDEKXXPKFLVIVAQKNHHTKFFQPGAPDNVPPGTVID 180
Query: 776 LGIM------IFIC---------------VLM------------LE*FLSYVYQRSTTAI 802
I ++C VL+ L LSYVYQRSTTAI
Sbjct: 181 NKICHPRNYDFYMCAHAGMIGTSRPTHYHVLLDEIGFSPDDLQELVHSLSYVYQRSTTAI 360
Query: 803 SVV 805
SVV
Sbjct: 361 SVV 369
>TC232387 similar to UP|AGO1_ARATH (O04379) Argonaute protein, partial (15%)
Length = 481
Score = 91.3 bits (225), Expect = 2e-18
Identities = 52/165 (31%), Positives = 91/165 (54%), Gaps = 7/165 (4%)
Frame = +3
Query: 229 VGGGVLSCKGLHSSFRTTQSGLSLNIDVSTTMIVRPGPVVDFLIENQN----VRDPFSLD 284
+G G+ S +G + S R TQ GLSLNID+S+T + P PV+DF+ + N R D
Sbjct: 12 LGEGLESWRGFYQSIRPTQMGLSLNIDMSSTAFIEPLPVIDFVTQLLNRDVSARPLSDAD 191
Query: 285 WNKAKRTLKNLRITAK---PSNQEYKITGLSELSCKDQLFTMKKRGAVAGEDDTEEITVY 341
K K+ L+ +++ ++Y+I+GL+ + ++ F + +RG + +V
Sbjct: 192 RVKIKKALRGIKVEVTHRGNMRRKYRISGLTSQATRELTFPVDERGTMK--------SVV 347
Query: 342 DYFVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSLISLQRYT 386
+YF +Q++ PC+ VG +RP Y+P+E+C ++ QRY+
Sbjct: 348 EYFYETYGFVIQHTQ-WPCLQVGNAQRPNYLPMEVCKIVEGQRYS 479
>AW460056
Length = 151
Score = 83.2 bits (204), Expect = 4e-16
Identities = 37/50 (74%), Positives = 43/50 (86%), Gaps = 1/50 (2%)
Frame = +2
Query: 439 TQVDGRVLQAPRLKFGN-EDFNPRNGRWNFNNKKFVEPVSLGNWSVVNFS 487
T+V+GRVLQAPRLKFGN EDFNPRNGRWNFNNKK V+P + W+ V+FS
Sbjct: 2 TEVEGRVLQAPRLKFGNGEDFNPRNGRWNFNNKKIVKPTKIERWAAVHFS 151
>AW755411 weakly similar to GP|20197419|gb| Argonaute (AGO1)-like protein
{Arabidopsis thaliana}, partial (4%)
Length = 416
Score = 78.6 bits (192), Expect = 1e-14
Identities = 39/72 (54%), Positives = 52/72 (72%), Gaps = 6/72 (8%)
Frame = +1
Query: 12 PPPPIVPPNVKPIKIEQ------EHLKKKLPTKAPMARRGLGTKGAKLPLLTNHFEVNVA 65
PPPP+VP ++ P+K E+ EH+KKK ++ P+AR GLG+KG K+ LLTNHF+VNVA
Sbjct: 199 PPPPVVPSDIVPLKAEEVLCTPTEHIKKKA-SRLPIARSGLGSKGNKIQLLTNHFKVNVA 375
Query: 66 NTNRVFFQYSVA 77
+ FF YSVA
Sbjct: 376 KNDGHFFHYSVA 411
>BG651396 similar to PIR|T01113|T011 translation initiation factor eIF-2C
homolog T21L14.12 - Arabidopsis thaliana, partial (5%)
Length = 273
Score = 69.3 bits (168), Expect(2) = 3e-14
Identities = 30/44 (68%), Positives = 37/44 (83%)
Frame = +2
Query: 558 NSDLYGPWKKKNLAEFGIVTQCIAPTRVNDQYLTNVLLKINAKL 601
+S GPWKKK L+E G+VTQCIAP ++ DQYLTNVLLKIN+K+
Sbjct: 128 HSSTLGPWKKKCLSEIGVVTQCIAPVKITDQYLTNVLLKINSKV 259
Score = 28.1 bits (61), Expect(2) = 3e-14
Identities = 10/14 (71%), Positives = 13/14 (92%)
Frame = +1
Query: 550 LCLLPERKNSDLYG 563
LC+LPERK+ D+YG
Sbjct: 22 LCVLPERKHCDIYG 63
>TC229952 homologue to UP|Q8LP00 (Q8LP00) ZLL/PNH homologous protein, partial
(20%)
Length = 913
Score = 63.9 bits (154), Expect = 3e-10
Identities = 34/77 (44%), Positives = 46/77 (59%)
Frame = +2
Query: 690 RELLLDFFHSSEERRPENIIIFRDGVSESQFNEVLNVELSQIIEACKFLDENWNPKFMVI 749
RELLL F ++ ++ P II +RDGVSE QF +VL EL I +AC L+ ++ P +
Sbjct: 65 RELLLSFKKATGQK-PLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEPSYQPPVTFV 241
Query: 750 VAQKNHHTKFFQPRSPD 766
V QK HHT+ F D
Sbjct: 242 VVQKRHHTRLFANNHDD 292
>BU926798 homologue to SP|Q9XGW1|PINH PINHEAD protein (ZWILLE protein).
[Mouse-ear cress] {Arabidopsis thaliana}, partial (14%)
Length = 446
Score = 63.2 bits (152), Expect = 5e-10
Identities = 36/138 (26%), Positives = 76/138 (54%), Gaps = 3/138 (2%)
Frame = +3
Query: 287 KAKRTLKNLRI--TAKPS-NQEYKITGLSELSCKDQLFTMKKRGAVAGEDDTEEITVYDY 343
K K+ L+ +++ T + S ++Y+++GL+ ++ +F + + + +V +Y
Sbjct: 60 KIKKALRGVKVEVTHRGSVRRKYRVSGLTSQPTRELVFPVDENSTMK--------SVVEY 215
Query: 344 FVHRRKIDLQYSAGLPCINVGKPKRPTYIPIELCSLISLQRYTKALSTSQRSSLVEKSRQ 403
F +QY+ LPC+ VG K+ Y+P+E C ++ QRYTK L+ Q ++L++ + Q
Sbjct: 216 FQEMYGFTIQYTH-LPCLQVGNQKKANYLPMEACKIVEGQRYTKRLNEKQITALLKVTCQ 392
Query: 404 KPVERMRVLSNALKASNY 421
+P +R + ++ + Y
Sbjct: 393 RPRDRENDILRTVQHNAY 446
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,607,186
Number of Sequences: 63676
Number of extensions: 504506
Number of successful extensions: 3981
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 3462
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3864
length of query: 826
length of database: 12,639,632
effective HSP length: 105
effective length of query: 721
effective length of database: 5,953,652
effective search space: 4292583092
effective search space used: 4292583092
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC124972.12


