

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124972.10 - phase: 1 /pseudo
(135 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
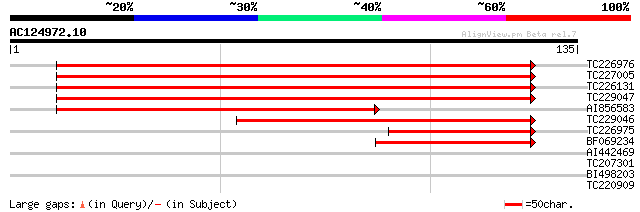
Score E
Sequences producing significant alignments: (bits) Value
TC226976 similar to UP|Q8S9L4 (Q8S9L4) At2g43020/MFL8.12, partia... 181 9e-47
TC227005 similar to UP|Q8S9L4 (Q8S9L4) At2g43020/MFL8.12, partia... 166 2e-42
TC226131 similar to UP|Q9SHX4 (Q9SHX4) F1E22.18, partial (86%) 142 4e-35
TC229047 weakly similar to GB|AAN40705.2|28933849|AF226656 perox... 137 1e-33
AI856583 92 1e-19
TC229046 similar to UP|Q9SHX4 (Q9SHX4) F1E22.18, partial (36%) 92 1e-19
TC226975 similar to UP|Q8S9L4 (Q8S9L4) At2g43020/MFL8.12, partia... 62 1e-10
BF069234 49 5e-07
AI442469 similar to GP|18377829|gb At1g62830/F23N19_19 {Arabidop... 30 0.26
TC207301 similar to UP|Q8RY08 (Q8RY08) AT5g59210/mnc17_100, part... 27 3.8
BI498203 26 6.5
TC220909 similar to UP|Q8RX84 (Q8RX84) AT5g28150/T24G3_80, parti... 25 8.5
>TC226976 similar to UP|Q8S9L4 (Q8S9L4) At2g43020/MFL8.12, partial (92%)
Length = 2336
Score = 181 bits (459), Expect = 9e-47
Identities = 90/114 (78%), Positives = 96/114 (83%)
Frame = +3
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L K IKFEPKL DWKEAAI+DI VG+ENKIILHFKNVFWPN +FL VVAE S GCSYFL
Sbjct: 1128 LKAKSIKFEPKLPDWKEAAISDIGVGIENKIILHFKNVFWPNVEFLGVVAETSYGCSYFL 1307
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA G VLVYMP G+ AKDIEKMSDEAAANFAF QLKKILPD SSP+ +V
Sbjct: 1308 NLHKAMGRPVLVYMPAGQLAKDIEKMSDEAAANFAFMQLKKILPDTSSPIQYLV 1469
>TC227005 similar to UP|Q8S9L4 (Q8S9L4) At2g43020/MFL8.12, partial (56%)
Length = 1094
Score = 166 bits (421), Expect = 2e-42
Identities = 81/114 (71%), Positives = 92/114 (80%)
Frame = +2
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L K I FEPKL DWKEAAIAD+ +G+ENKIILHF+NVFWPN +FL VVA+ CSYFL
Sbjct: 197 LKAKKILFEPKLPDWKEAAIADLGIGLENKIILHFENVFWPNVEFLGVVADTPYECSYFL 376
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA G +VLVYMP G+ AKD+EKM DEAA NFAF QLKKI PDASSP+ +V
Sbjct: 377 NLHKATGRAVLVYMPSGQLAKDVEKMPDEAAVNFAFMQLKKIFPDASSPIQYLV 538
>TC226131 similar to UP|Q9SHX4 (Q9SHX4) F1E22.18, partial (86%)
Length = 2250
Score = 142 bits (359), Expect = 4e-35
Identities = 72/114 (63%), Positives = 82/114 (71%)
Frame = +2
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L LI+FEPKL DWK +AI+D+ VG ENKI L F VFWPN + L VA S C YFL
Sbjct: 920 LKANLIEFEPKLPDWKVSAISDLGVGNENKIALRFDKVFWPNVELLGTVAPTSYTCGYFL 1099
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA GH VLVYM GR A DIEK+SDEAAANF +QLKK+ P+AS PV +V
Sbjct: 1100 NLHKATGHPVLVYMVAGRFAYDIEKLSDEAAANFVMQQLKKMFPNASKPVQYLV 1261
>TC229047 weakly similar to GB|AAN40705.2|28933849|AF226656 peroxisomal
N1-acetyl-spermine/spermidine oxidase {Mus musculus;} ,
partial (7%)
Length = 706
Score = 137 bits (346), Expect = 1e-33
Identities = 68/114 (59%), Positives = 80/114 (69%)
Frame = +1
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L LI+F PKL WK AI DI +G ENKI L F VFWPN + L +VA S C YFL
Sbjct: 271 LKANLIEFSPKLPHWKAEAIKDIGMGNENKIALRFDAVFWPNVEVLGIVAPTSYACGYFL 450
Query: 72 NLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
NLHKA GH +LVYM G+ A D+EK+SDE+AANFA +QLKK+ PDAS PV +V
Sbjct: 451 NLHKATGHPILVYMAAGKFAYDLEKLSDESAANFAMQQLKKMFPDASKPVQYLV 612
>AI856583
Length = 296
Score = 91.7 bits (226), Expect = 1e-19
Identities = 45/77 (58%), Positives = 50/77 (64%)
Frame = +1
Query: 12 LLTKLIKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPNEDFLEVVAEISNGCSYFL 71
L LI+F PKL WK AI DI +G ENKI L F VFWPN + L +VA S C YFL
Sbjct: 22 LKANLIEFSPKLPHWKAEAIKDIGMGNENKIALRFDAVFWPNVEVLGIVAPTSYACGYFL 201
Query: 72 NLHKAAGHSVLVYMPVG 88
NLHKA GH +LVYM G
Sbjct: 202NLHKATGHPILVYMAKG 252
>TC229046 similar to UP|Q9SHX4 (Q9SHX4) F1E22.18, partial (36%)
Length = 893
Score = 91.7 bits (226), Expect = 1e-19
Identities = 43/71 (60%), Positives = 53/71 (74%)
Frame = +1
Query: 55 DFLEVVAEISNGCSYFLNLHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKIL 114
+ L +VA S C YFLNLHKA GH +LVYM G+ A D+EK+SDE+AANFA +QLKK+
Sbjct: 226 EVLGIVAPTSYACGYFLNLHKATGHPILVYMAAGKFAYDLEKLSDESAANFAMQQLKKMF 405
Query: 115 PDASSPVTIIV 125
PDAS PV +V
Sbjct: 406 PDASKPVQYLV 438
>TC226975 similar to UP|Q8S9L4 (Q8S9L4) At2g43020/MFL8.12, partial (24%)
Length = 979
Score = 61.6 bits (148), Expect = 1e-10
Identities = 30/35 (85%), Positives = 32/35 (90%)
Frame = +3
Query: 91 AKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
AKDIEKMSDEAAANFAF QLKKILPDASSP+ +V
Sbjct: 6 AKDIEKMSDEAAANFAFMQLKKILPDASSPIQYLV 110
>BF069234
Length = 349
Score = 49.3 bits (116), Expect = 5e-07
Identities = 23/38 (60%), Positives = 29/38 (75%)
Frame = +1
Query: 88 GRQAKDIEKMSDEAAANFAFKQLKKILPDASSPVTIIV 125
GR DIEK+SDEAAANF +QLKK+ P++S PV +V
Sbjct: 1 GRFGYDIEKLSDEAAANFVMQQLKKMFPNSSKPVQYLV 114
>AI442469 similar to GP|18377829|gb At1g62830/F23N19_19 {Arabidopsis
thaliana}, partial (12%)
Length = 334
Score = 30.4 bits (67), Expect = 0.26
Identities = 27/101 (26%), Positives = 44/101 (42%), Gaps = 4/101 (3%)
Frame = +1
Query: 17 IKFEPKLLDWKEAAIADIRVGVENKIILHFKNVFWPN--EDFLEVVAEISNGCSYFL--N 72
I+F P+L K+ AI + G+ NK+ + F FW + F + ++S +FL +
Sbjct: 28 IEFVPELPQRKKDAIHRLGFGLLNKVAILFPYNFWGGDIDTFGHLTEDLSMRGEFFLFYS 207
Query: 73 LHKAAGHSVLVYMPVGRQAKDIEKMSDEAAANFAFKQLKKI 113
+G +LV + A E MS + LK I
Sbjct: 208 YSSVSGGPLLVALVAWEAAIXFEMMSPVESVKRVLDILKNI 330
>TC207301 similar to UP|Q8RY08 (Q8RY08) AT5g59210/mnc17_100, partial (39%)
Length = 1189
Score = 26.6 bits (57), Expect = 3.8
Identities = 9/36 (25%), Positives = 22/36 (61%)
Frame = +2
Query: 63 ISNGCSYFLNLHKAAGHSVLVYMPVGRQAKDIEKMS 98
+ CS+ LNL+K+ H ++++ +G+ ++ +S
Sbjct: 767 LQKSCSFLLNLNKSNKHMMVLHASIGQCCATLKAIS 874
>BI498203
Length = 338
Score = 25.8 bits (55), Expect = 6.5
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -2
Query: 49 VFWPNEDFLEVVAE-----ISNGCSYFLNLHKAAGHSVLVYMPVG 88
VFWPN+ V + N C F +L A HS+L+ +P G
Sbjct: 229 VFWPNQHQSPVRLFWPHYWLKNTCH*FPSLVGAVLHSMLLLLPAG 95
>TC220909 similar to UP|Q8RX84 (Q8RX84) AT5g28150/T24G3_80, partial (22%)
Length = 782
Score = 25.4 bits (54), Expect = 8.5
Identities = 11/30 (36%), Positives = 17/30 (56%)
Frame = +2
Query: 10 ARLLTKLIKFEPKLLDWKEAAIADIRVGVE 39
+RLLT I+ E +L WK + D+ V +
Sbjct: 425 SRLLTDFIRVEERLELWKSTILTDLLVSCD 514
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,669,088
Number of Sequences: 63676
Number of extensions: 65423
Number of successful extensions: 420
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 420
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 420
length of query: 135
length of database: 12,639,632
effective HSP length: 87
effective length of query: 48
effective length of database: 7,099,820
effective search space: 340791360
effective search space used: 340791360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 53 (25.0 bits)
Medicago: description of AC124972.10


